Transposons families/Tn3 family
Contents
- 1 Historical
- 2 General Organization
- 2.1 Diversity: TnpA Tree
- 2.2 Tn3 family complementation groups
- 2.3 Tn3 and Tn21 groups
- 2.3.1 The Tn21 Clade
- 2.3.1.1 Derivatives with a simple mercury operon.
- 2.3.1.2 Derivatives with class I integrons: 2 events leading to multiple antibiotic resistance
- 2.3.1.3 Derivatives with upstream passenger genes: colistin resistance.
- 2.3.1.4 Derivatives with upstream passenger genes: other passengers.
- 2.3.1.5 Derivatives with divergent tnpR and tnpA
- 2.3.2 The Tn21 Lineage.
- 2.3.3 Tn1721 and (tandem) amplification of the tet genes
- 2.3.4 The Tn163 Clade
- 2.3.5 The Tn4430 Clade
- 2.3.6 The Tn3 Clade
- 2.3.7 The Tn3000 Clade
- 2.3.8 The Tn4651 Clade
- 2.3.9 The Tn1071 Clade
- 2.3.1 The Tn21 Clade
- 3 MITES, MICs and TALES
- 4 Acquisition of Passenger Genes.
- 5 Transposition Mechanism Overview
- 5.1 Early Studies
- 5.2 Replicative transposition
- 5.3 Interaction of transposase and transposon ends
- 5.4 TnpA functional domains
- 5.5 Cleavage and Strand transfer.
- 5.6 Mechanism in the Light of Structure
- 5.7 Tn3 Transposition immunity, a poorly understood phenomenon.
- 5.8 On Ended Transposition.
- 5.9 Resolution
- 5.9.1 The serine recombinases.
- 5.9.2 Studies with Tn1000 (γδ) and Tn3 res.
- 5.9.3 Tn3 res, tnpR and tnpA gene expression.
- 5.9.4 The Mechanics of Resolution.
- 5.9.5 The Tn3 synaptosome
- 5.9.6 The Tn1721, Tn21 and Tn501 res.
- 5.9.7 Tn res activity tnpR and tnpA gene expression.
- 5.9.8 The long serine recombinases
- 5.9.9 Serine-recombinases which use IHF/Hu: the Sin Synaptosome.
- 5.9.10 The irs/TnpI system
- 5.9.11 The Mechanics of Resolution.
- 5.9.12 Irs, tnpR and tnpA and gene expression.
- 5.9.13 The rst/TnpS/T system.
- 5.10 Toxin-Antitoxin genes: Special Passengers linked to the transposition process?
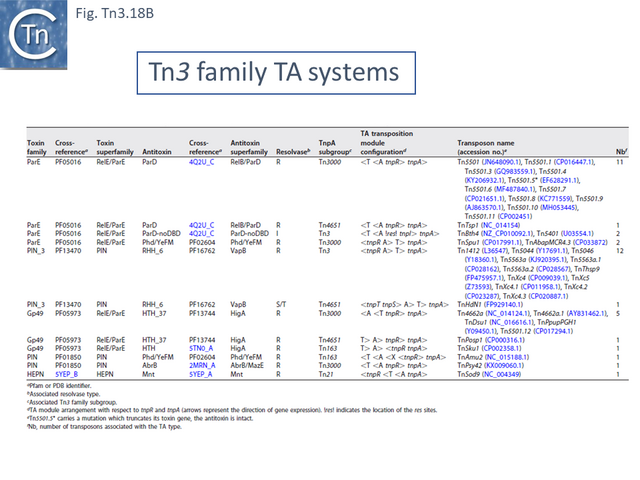
- 5.10.1 Identification of TA gene pairs in Tn3 family members.
- 5.10.2 TA diversity in Tn3 family members.
- 5.10.3 TA distribution and organization within the Tn3 family
- 5.10.4 Acquisition and exchange of TA modules.
- 5.10.5 Tn3 family-associated TA passenger gene are located in a unique position.
- 5.10.6 Regulation of Tn3 family TA gene expression.
- 5.10.7 Tn3 family with TnpR and TnpRL
- 5.10.8 Tn3 family with TnpI
- 5.10.9 Tn3 family with TnpS/T
- 5.10.10 A model for T/A activity in transposon transposition.
- 6 Conclusion and Future.
- 7 Bibliography
Historical
Members of the Tn3 family were among the earliest transposons to be identified. In fact, the word “transposon” was used for the first time in 1974 by Hedges and Jacob in a seminal article in which they showed that ampicillin resistance could be transmitted between a number of different plasmids [1]:
“We designate DNA sequences with transposition potential as transposons (units of transposition) and the transposon marked by the ampicillin resistance gene(s) as transposon A “.
TnA, later called Tn1, was isolated from the plasmid RP4 [1] while the closely related TnB and TnC (later called Tn2 and Tn3 respectively) were isolated from plasmids RSF1010 [2] and R1 [3][4]. Tn3 proved to be inserted into another, larger Tn3 family transposon, Tn4 [4]. A number of early studies using electron microscope DNA heteroduplex analysis (e.g. [5][6][7] Fig. Tn3.1) demonstrated that movement of ampicillin resistance was accompanied by insertion of a DNA segment of about 4-5 kilobases (kb).
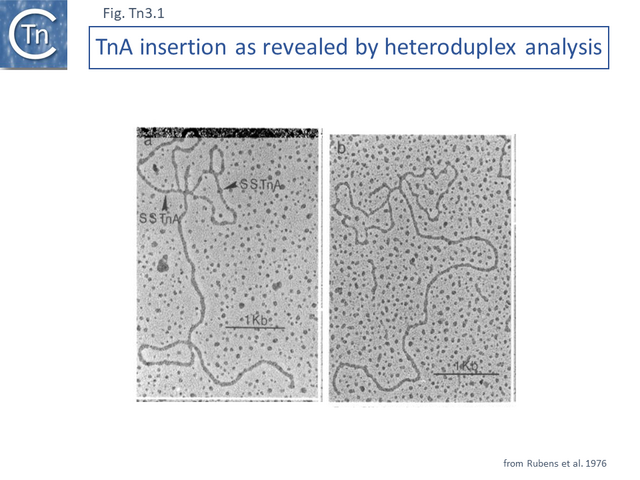
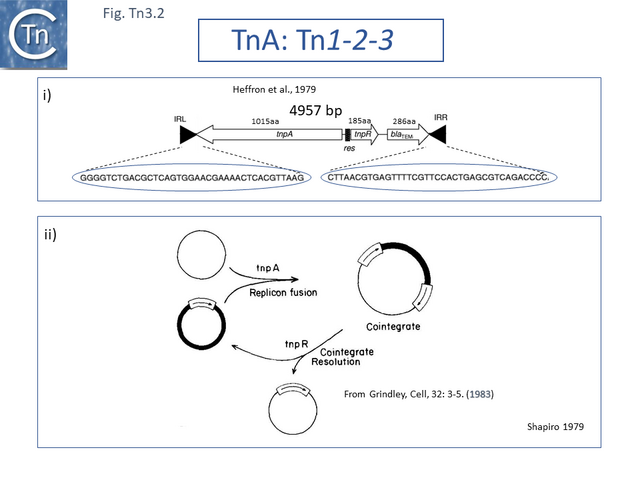
The DNA sequence of the 4957 base pair (bp) Tn3 was obtained in 1979 [8] and shown to be bordered by two inverted repeat sequences of 38 bp and included 2 genes in addition to the ampicillin resistance (beta-lactamase, bla) gene: a transposase gene, tnpA, and a gene involved in regulating tnpA and its own expression, tnpR (R for repressor). TnpR was subsequently shown to be a site-specific recombinase intimately involved in the transposition pathway [9] which acts on a specific site, IRS (Internal Resolution Site) (Fig. Tn3.2 i). In its absence, insertion of two complete, directly repeated, Tn3 copies occurred [8].
It was suggested that this type of structure was an intermediate in Tn3 transposition and that the IRS site was required for recombination and subsequent segregation of the direct repeats to leave a single copy of Tn3 [10] according to the Shapiro cointegrate model of replicative transposition (Fig. Tn3.2 ii; [11] Fig. 2.7 Early models).
Indeed, Tn3 was shown to be instrumental in permitting transfer of a non-transmissible plasmid by a co-resident conjugative plasmid [12] resulting in fusion of the two plasmids which were separated at their junctions by two directly repeated Tn copies [12][13][14][15].
A related TE, or Tn1000, was identified as part of the plasmid F and appeared as an insertion loop in heteroduplex analysis [15][17]. It was also implicated in the integration of the F plasmid into the Escherichia coli host chromosome [17] and deletion of chromosomal DNA in F’ plasmids [18][19] derived from F-excision with flanking chromosomal DNA [20]. It generates 5bp direct target repeat (DR) on insertion [21] and carries similar ends to those of Tn3 and to IS101, a small 200bp sequence carried by the pSC101 plasmid [22][23].
Many other related transposons have since been identified with a highly diverse range of passenger genes (see [24] and Fig. Tn3.4B). The tetracycline resistance transposon, Tn1721 from plasmid pRSD1 [25] and the multi-resistance transposons, Tn4 from R6-5 and Tn21, a component of the 25 kb resistance determinant (r-det) of the plasmid NR1 (R100) [6] are two of many early examples.
General Organization
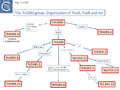
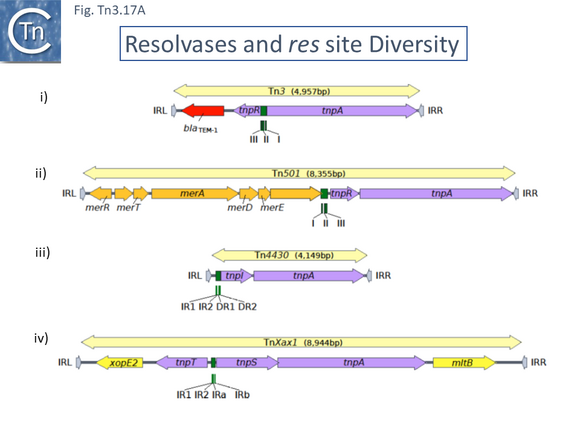
Members of the Tn3 transposon family form a tightly knit group with related transposases and DNA sequences at their ends. The basic Tn3 family transposition module is composed of transposase and resolvase genes and two ends with related terminal inverted repeat DNA sequences, the IRs, of 38-40bp or sometimes even longer (Fig. 3.2 i) [26].
There is a large (~1000 aa) DDE transposase, TnpA, significantly longer than the DDE transposases normally associated with Insertion Sequences (IS) (see [27]). TnpA catalyzes the DNA cleavage and strand transfer reactions necessary for formation of a cointegrate transposition intermediate during replicative transposition.
A second feature of members of this transposon family is that they carry short (~100-150bp) DNA segments, res (for resolution) or rst (for resolution site tnpS tnpT – see below; [28]) at which site-specific recombination between each of the two Tn copies occurs to “resolve” the cointegrate into individual copies of the transposon donor and the target molecules each containing a single transposon copy (Fig. Tn3.2 ii)(see [24]). This highly efficient recombination system is assured by a transposon-specified sequence-specific recombinase enzyme: the resolvase.
There are at present three known major resolvase types: TnpR (which includes two subgroups, long and short with and without a C-terminal extension; Resolution), TnpI, and TnpS+TnpT, distinguished, among other things, by the catalytic nucleophile involved in DNA phosphate bond cleavage and rejoining during recombination: TnpR, a classic serine (S)-site-specific recombinase (e.g. [29][30]); TnpI, a tyrosine (Y) recombinase similar to phage integrases [31] (see [24]); and a heteromeric resolvase combining a tyrosine recombinase, TnpS, and a divergently expressed helper protein, TnpT, with no apparent homology to other proteins [28][32].
The resolvase genes can be either co-linear, generally upstream of tnpA or divergent. In the former case the res site lies upstream of tnpR and in the latter case, between the divergent tnpR and tnpA genes. For relatives encoding TnpS and TnpT, the corresponding genes are divergent and the res (rst) site lies between tnpS and tnpT.
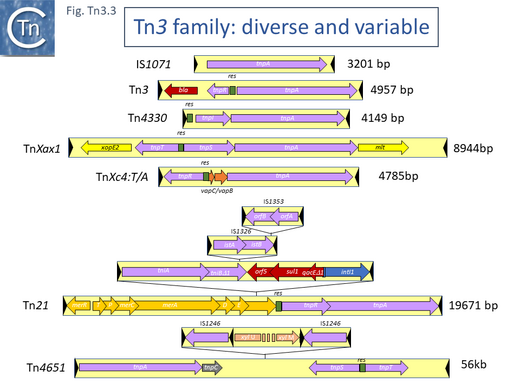
Examples of these architectures are shown in Fig. Tn3.3. Each res includes a number of short DNA sub-sequences which are recognized and bound by the cognate resolvases. These are different for different resolvase systems. But where analyzed, res sites also include promoters which drive both transposase and resolvase expression. Indeed, TnpR from Tn3 was originally named for its ability to repress transposase expression by binding to these sites [8][10] (see later: Tn3 family resolution systems).
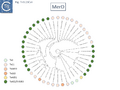
Diversity: TnpA Tree
The complexity of these Tn resides in the diversity of other mobile elements incorporated into their structures (such as IS and integrons as well as other Tn3 family members – see [24] - and other passenger genes). The most notorious of these genes are those for antibiotic and heavy metal resistance although other genes involved in organic catabolite degradation and virulence functions for both animals and plants (Fig. Tn3.3) also form part of the Tn3 family arsenal of passenger genes.
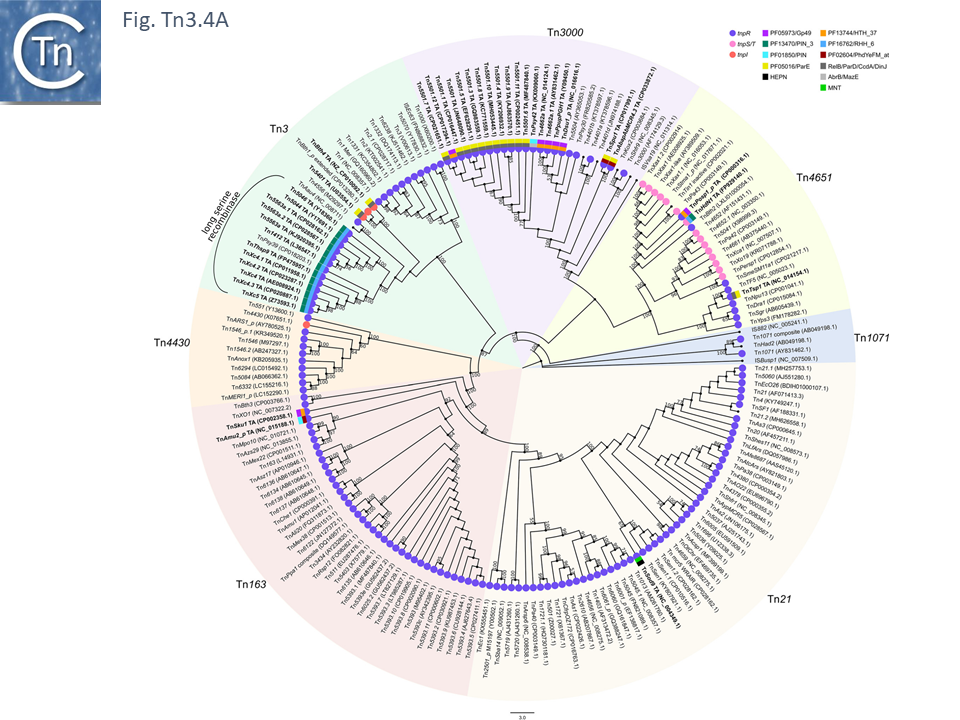
The diversity of Tn3 family members was investigated using a library of carefully annotated examples in the ISfinder database [33], those listed in Nicolas et al. [24], those resulting from a search of NCBI for previously annotated Tn3 family members (March 2018) and those obtained using a script, Tn3_TA_finder, which can searched for tnpA, tnpR, genes located in proximity to each other (Tn3finder, https://tncentral.proteininformationresource.org/TnFinder.html; Tn3_TA_finder, https://github.com/danillo-alvarenga/tn3-ta_finder) in complete bacterial genomes in the RefSeq database at NCBI. This yielded 190 Tn3 family transposons for which relatively complete sequence data (transposase, resolvase, and generally both IRs) were available.
Full annotations can be found at TnCentral (https://tncentral.proteininformationresource.org/index.html). A tree based on the transposases of these transposons is shown in Fig. Tn3.4A [34].
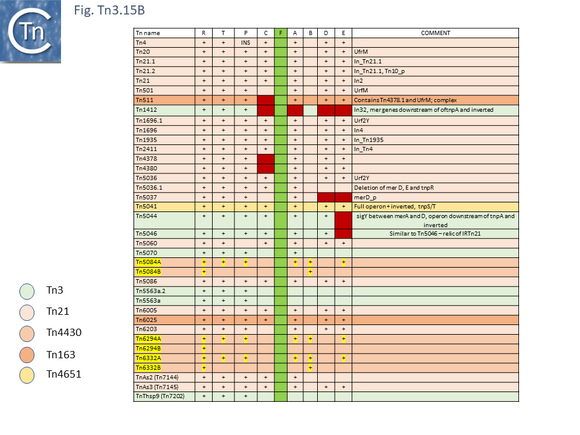
The tree defines 7 deeply branching clades which supports the divisions proposed by Nicolas et al., [24]. They were named after a representative Tn from each clade: Tn3; Tn4651; Tn3000; Tn1071; Tn21; Tn163; and Tn4330. As can be seen from Fig. Tn3.4A, the vast majority of Tn3 family members encode a tnpR/res resolution system and encode a TnpR without the C-terminal extension (shown by blue circles) and a small group which encodes a TnpR derivative with the C-terminal extension (Fig. Tn3.4A). However, a significant sub-group of the Tn4651 clade encodes the tnpS/tnpT/rst resolution system (pink circles) while the tnpI/irs is represented in only three cases.
An overview, extracted from TnCentral, of the diversity and distribution of different passenger genes within the Tn3 family and their presence in different bacterial hosts is shown in Fig. Tn3.4B.
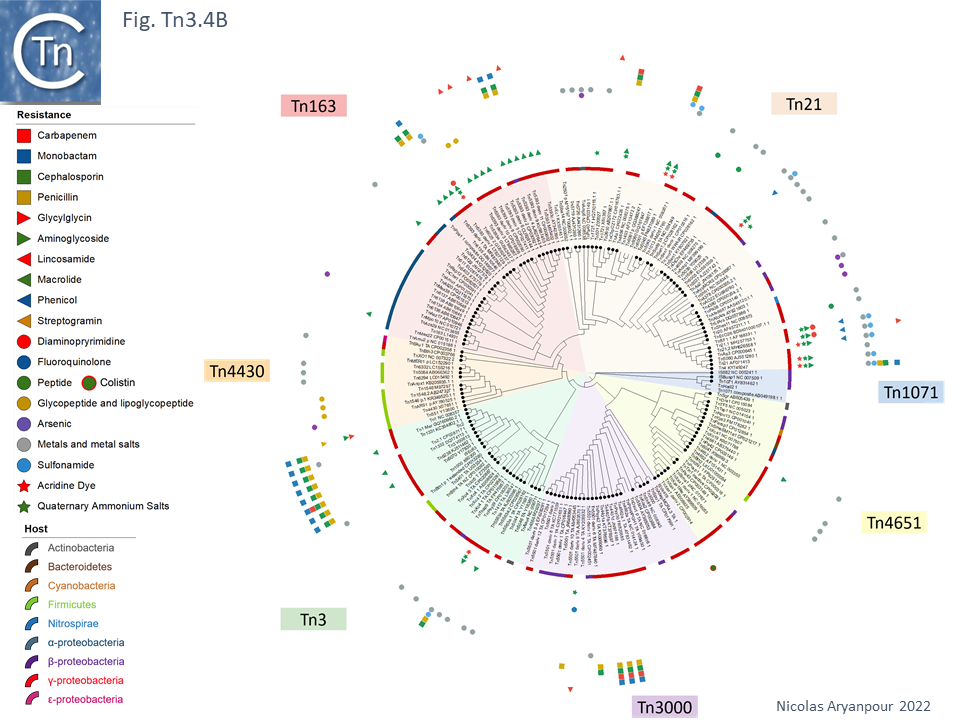
Tn3 family complementation groups
Early studies on the relationship between different Tn3 family members revealed that they could be divided into different functional groups by genetic complementation of their tnpA and tnpR genes [35][36].
Transposition-deficient tnpA mutants of Tn1721 (Tn21 clade; Fig. Tn3.4A) and the mercury resistance transposon Tn501 [37][38][39][40] (close to Tn1721 in the Tn21 clade;) could be complemented in trans by co-resident wild type copies of either Tn21, Tn501, or Tn1721, while transposition of a Tn21 tnpA mutant could only be restored by Tn21. Moreover, Tn3 was unable to complement either Tn21, Tn501, or Tn1721, and vice versa [36]. Similarly, a Tn21 tnpR mutant could be complemented by Tn21, Tn501 or Tn1721, but not by Tn3. Moreover, mutations in the Tn2603 tnpA and tnpR genes could be complemented by mercury resistance transposons Tn2613 and Tn501 (although Tn501 was much less efficient in complementation than Tn2613) but not by gamma delta, Tn2601 or Tn2602 (both of which resemble the Tn3 group – see Fig. Tn3.7 A) [41]. In this context, it is perhaps useful to note the Tn501 and Tn1721 are located at some distance from Tn21 in the tnpA phylogenetic tree. This reinforced the idea, based principally on the direction of transcription of their tnpA and tnpR genes, that the Tn3 family could be divided into 2 major groups: Tn3 and Tn21 [42].
Tn3 and Tn21 groups
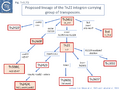
Grinsted et al. [43] identified at least five Tn3 family subgroups which correspond to those shown in Fig. Tn3.4A. In addition to the Tn3 and Tn21 subgroups, the others included Tn2501 (Tn163 subgroup), Tn917/Tn551 (Tn4430 subgroup) and Tn4556 (Tn3000 subgroup). Tn917 and Tn551 are quasi-identical and Tn4430 was included in a separate subgroup because it encodes a resI/tnpI resolution system.
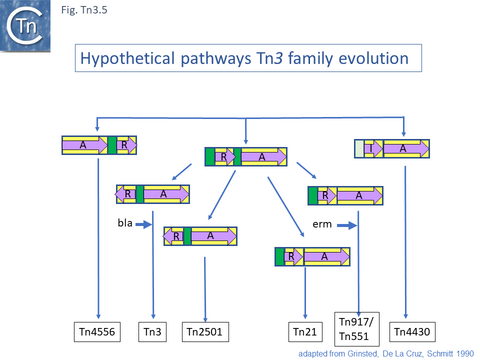
These divisions were based on the observations that: transposition proteins within each group were at least 70% similar or identical whereas this value was only about 30% between groups and that the IR sequences were less than 26/38 identical. The authors propose a model for the evolution of the Tn3 family transposition modules (Fig. Tn3.5) in which two ancestral modules were assembled: the first included a tnpR gene (which they suggest was flanked by an invertible DNA segment incorporating the res site) and a tnpA gene. This subsequently gave rise to each of the Tn3 subgroups by tnpR/res inversion and sequence divergence. For Tn such as Tn4430, the assembly involved tnpI/res and tnpA components. The tnpS/tnpT/rsc resolution system was not included since it had not been identified at that date but could easily be incorporated into this scheme. To our knowledge, the proposed ancestral components in this scheme have not yet been identified.
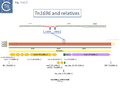
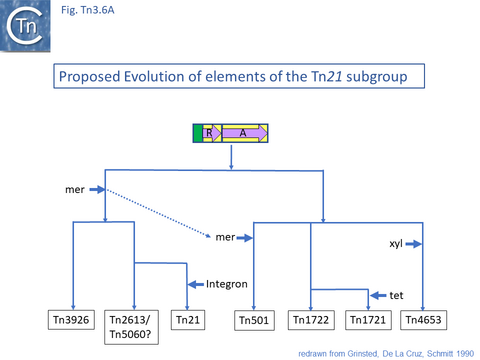
The diversification of different Tn21 clade members was also examined [43] (Fig. Tn3.6) and forms two subclades. One includes Tn21, Tn2613 (whose sequence is not available but which may be identical to Tn5060-AJ551280.1) and Tn3926 (with only a partial sequence available but which complements a tnpA-defective Tn21 but not Tn1721 or Tn501 mutants [44]). The other includes Tn501, Tn1722, Tn1721 and Tn4653. Tn501 and Tn1721 are located in a sub-clade distinct from Tn501 and Tn5060 (Fig. Tn3.4A). In this scheme, mercury resistance was proposed to have been acquired twice independently in each subclade, early in the Tn21 subclade lineage and later in the line leading to Tn501. The ancestor of Tn21 had acquired an integron platform transported by a Tn402 family transposon and Tn1721 was derived from Tn1722 by acquisition of a tet resistance gene.
The Tn21 Clade
The Tn21 is a large group with 49 members at present in TnCentral (most of these are shown in Fig. Tn3.7 A). Like the entire Tn3 family, Tn21 clade members possess highly conserved IRL and IRR (Fig. Tn3.7 B, C and D).
Many clade members encode tnpR with a res site immediately upstream and, in a majority (but not all), tnpA is located downstream and in the same orientation. The res sites of this class (Fig. Tn3.7 E) show a high degree of identity (Fig. Tn3.7 F). However other tnpR/tnpA configurations also occur (Fig. Tn3.3; Fig. Tn3.7 E) and their res sites (see below: The Tn1721, Tn21 and Tn501 res) show relatively good conservation (Fig. Tn3.7 F)
Derivatives with a simple mercury operon.
In general, passenger genes in this clade are located upstream of tnpR and the res site (Figs. Tn3.7G-N). Ten carry only genes for resistance to mercury salts.
Two of these, Tn5060 (AJ551280.1) (Tn3.7G), the proposed ancestor of the Tn21 integron group (Tn3.7I) [45], and Tn20 (AF457211.1) are nearly identical except for a few SNP and a deletion of a few base pairs in ufrM (Tn20).
These are quite different in sequence both in the mer operon and in tnpR/tnpA segments from the other transposons of similar organization. Tn1696.1 (CP047309) and Tn5036 (Y09025) differ by only a few SNPs while Tn4378 (CP000355), Tn6203 (CP065412) and Tn6346 (KM659090) are also quite different from the these. Tn4378 and Tn6203 show many sequence differences along their entire length as does TnAs2 (JN106175.1) while clearly, Tn6346 shares identity with Tn4378 over the entire length of the mer operon up to res but shows variability in the tnpR/tnpA region.
This clearly indicated that there has been an exchange by inter res recombination between two different transposons (Tn3.7H). A similar recombination has occurred with Tn501. In addition, Tn4380 appears to have been derived from Tn6346 by deletion of the entire res site. Thus Tn4378, Tn6436 (Tn4380) and Tn501 share highly related mer operons but vary in the sequences of tnpR and tnpA.
Derivatives with class I integrons: 2 events leading to multiple antibiotic resistance
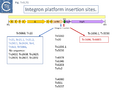
At least 22 Tn21 clade members carry class I integrons (Fig. Tn3.7 A and Fig. Tn3.7 I) although the DNA sequence of some of these is not available. These are transmitted by Tn402 derivative transposons which exhibit pronounced target specificity (more details at: Tn402 family) and show a preference for insertion into or close to Tn3 family res sites or into plasmid res sites. A major pathway for the acquisition of passenger genes was the initial integration of a Tn402-like transposon which carried a class I integron platform. The integron insertions have occurred at one of two positions in the Tn5060 /Tn20 related examples (Fig. Tn3.7 G). In one group, which all encode an identical mer operon, insertion occurred in a precise position in a gene of unknown function, ufrM (see: The Tn21 Lineage) (Fig. Tn3.7 I). Since these occur at the same nucleotide, it seems possible that all diverged from a single insertion event.
In the others, the res site itself has been targeted: at two slightly different positions both in the Tn1696 (Fig. Tn3.7 J) (also carrying a mer operon) and Tn1721 (with an mcp gene) groups (Fig. Tn3.7 K) while a third example can be observed in Tn5045.1 carrying the tao gene cluster (Fig. Tn3.7 L). The fact that integrons In2 and In4 are located in different sequence environments in two distinct mercury resistance transposons, Tn21 and Tn1696 has previously been noted [46].
Thus, although widespread in nature, class 1 integrons appear to have inserted in only six target sequences in the entire Tn21 clade in TnCentral. The significant variability therefore arises principally by acquisition and loss of integron cassettes and by frequent various degrees of loss by deletion/inactivation (see: Tn21 lineage) of the Tn401 transposition genes tniA,B,Q and its resolvase tniR (see: Tn402 family).
Derivatives with upstream passenger genes: colistin resistance.
Of the four colistin resistant examples (Fig. Tn3.7 M): TnSen1.1 [Tn7191] and TnSen1.2 [Tn7192] are nearly identical except that TnSen1.2 carries an ISPa96 insertion; both TnEc026 [Tn7159] and TnMCR5ECO26H11 [Tn7163] are identical but TnEcO26 has two right ends.
Moreover, while the left segment of all 4 are closely related, there appears to have been a recombination event in the region of the res site two right ends and TnSen11.2/TnSen1.2 and TnEcO26/ TnMCR5ECO26H11 carry divergent tnpR and tnpA.
Derivatives with upstream passenger genes: other passengers.
There are a number of other Tn21 clade members with different upstream passenger genes. Analysis of these reveals that, although there has been some diversification of the tnpR and tnpA genes (Fig. Tn3.7 N), there is a clear breakpoint in identity which occurs at the res site. Sequence analysis (Fig. Tn3.7 N) indicates that the break in identity occurs at the potential AT recombination dinucleotide (see: Resolution topic below) strongly suggesting that acquisition of various passenger genes frequently occurs by modular exchange via inter-res recombination.
Derivatives with divergent tnpR and tnpA
There are a number of Tn21 clade members in which the tnpR and tnpA genes are expressed divergently. Several of these (e.g. Tn4659, TnAcsp1 [Tn7133], TnEc1 [Tn7158] and TnSba14 [Tn7190]) (Fig. Tn3.7 O) do not encode passenger genes and are not closely related, while others encode heavy metal resistance operons located between tnpR and tnpA (e.g. TnLfArs [Tn7162], TnOtChr [Tn7169]) while TnPa38 [Tn7172] encodes genes of unknown function and TnSod9 [Tn7199] is the only example in the Tn21 clade to encode a Toxin/Antitoxin gene pair. These are not closely related.
The Tn21 Lineage.
The Tn21 lineage is an example of the plasticity of Tn3 family transposons. Tn21 was originally identified in the multiple antibiotic resistance plasmid NR1/R100 [56], as part of the IS1-flanked r-determinant [5] and its component antibiotic resistance genes were first mapped by restriction enzyme digestion and cloning [57]. The Tn21 group of transposons appear to be very successful as judged by their distribution.
This is arguably the result of acquisition of an integron platform permitting incorporation of various resistance genes as integron cassettes [43][58] (Fig. Tn3.7 A and Fig. Tn3.7 G). Tanaka and collaborators proposed in the early 1980s that Tn21-like transposons which carry a variety of antibiotic resistance genes are related and evolved from an ancestor carrying a mercury resistance operon [59] (Fig. Tn3.5; Fig. Tn3.7 P).
Tn21 itself is a complex collection of intercalated TE and a comprehensive and detailed schemes for its formation has been proposed [43][58][59] (see Fig. Tn3.6.; Fig. Tn3.7 P). Unfortunately, although the DNA sequences of some of the component transposons are now available (e.g. Tn4, Tn21, Tn2411), many are not and comparison was based on physical and functional maps (restriction, genetic features) [41][59][60][61].
This scheme was later expanded with the addition of more up-to-date information to include a number of potential Tn21 descendants (see [58]) (Fig. Tn3.7 Q). It was proposed that a Tn21 precursor (Tn21) acquired an integron platform such as is found in Tn4 (for convenience, called In_Tn4 here) which then received an insertion of IS1353 into a resident IS1326 copy to generate In2 found in Tn21 [59].
Although the Tn21 group ancestor prior to acquisition of the mercury resistance genes is at present unknown, the later identification of a mercury resistance transposon, Tn5060 (AJ551280.1), isolated from the Siberian permafrost [45] (Fig. Tn3.7 R) provided a possible candidate for the hypothetical Tn21 precursor, Tn21°.
Other examples of this Tn such as Tn20 (AF457211) (Fig. Tn3.7 I) can be identified which share a number snips with other members of the group compared to Tn5060 [62] and therefore is perhaps a better candidate as an ancestor.
An alternate view of the path from Tn5060 to Tn21 is that evolution of the integron platform occurred “in situ” by the gradual loss/accumulation of component TE. In this scheme (Fig. Tn3.7 S and Fig. Tn3.7 P), a first step would be insertion into the ufrM (unknown function) gene of a Tn402 family transposon to provide the integron platform (Fig. Tn3.7 S).
Although it has been shown that transposition of defective Tn402 transposons (e.g. In0 and In2) can be complemented by a related, wildtype copy [63], it seems simpler to hypothesize that an initial insertion involved a Tn402 derivative with a complete functional set of Tn402 transposition genes. We have chosen a simple integron platform, In_Tn1721.1 from Tn1721.1 (HQ730118.1), for convenience.
This carries tniA,B,Q, the resolvase tniR together with the Tn402 res site, both ends (IRt and IRi), the integron integrase int and a common qac gene cassette. Insertion into the Tn5060 urfM gene generates a 5 bp DR (Fig. Tn3.7 S) and leads to the formation of tnpM from the 3’ end of ufrM (serendipitously generating an ATG initiation codon) [58][64]. TnpM has been suggested to be a transposition regulatory gene (but see Resolution below). Subsequent steps in the Tn21 lineage (Fig. Tn3.7 T) would then involve modification of the integron platform by acquisition of the typical GNAT (previously known as orf5) and sul genes, decay of the Tn402 transposition genes and insertion, first of IS1326 (resulting in In0) followed by acquisition of the aadA integron cassette (generating In_Tn4) and, finally, insertion of IS1353 into IS1326 (IS1326::IS1353) between IRL and the start of the istA gene presumably not affecting IS1326 transposition functions (generating In2).
Due to their conservation in a large number of class I integron platforms, the DNA region including the sul, qac and GNAT family (previously called orf5) genes has been called the 3’CS (conserved segment) while that including the attI site and intI gene has been called the 5’CS [65] (however, using a more extended data set it was noted that, while the 5’CS was highly conserved across a number of integrons, the 3’CS proved to be somewhat more variable [66]).
Tn2411 is not only the precursor of Tn21. It was proposed to give rise to additional transposons (Fig. Tn3.7 Q)[58]: to Tn4 by insertion of a Tn3 transposon copy into the merP gene (Fig. Tn3.7 U); to Tn5086 [67] by deletion of the In_Tn4 IS1326 copy to generate Tn2608 [58] and replacement of the aadA cassette and acquisition of dfrA7 (Fig. Tn3.7 V); and to Tn2410 by replacement of the aadA cassette by an oxa cassette [61].
The complete DNA sequences of many of these Tn are not available but Tn5086 or Tn2608 could be reconstructed from Tn21 using the limited sequence data in refeference [67]. Moreover, using the reconstructed Tn5086 sequence in a BLAST search revealed an identical sequence in the E. coli SCU-164 chromosome (CP054343) and a nearly identical copy, in which the IRL had been interrupted by an insertion of IS4321, in E. coli plasmid pSCU-397-2 (CP054830) in addition to many closely related copies.
This analysis suggests that deletion of IS1326 had occurred by nearly-precise excision [68] since the deletion junction observed in Tn5086 [67] is not the original sequence identified in Tn2411. Indeed, the DNA sequences of Tn2411, Tn2608 and Tn5086, (Fig. Tn3.7 V) suggest that In_Tn2608 and In22 were derived by deletion from a structure similar to In_Tn4 because neither carry an IS1326 copy although they both retain the tip of the IRL (4 bp for In_Tn2608 and 3bp for In22) at one end and are missing 5bp of In_Tn4 DNA flanking the right IS1326 end.
Tn21 was also proposed to give rise to a number of different transposons [58][61]: to Tn1831 by IS1326-mediated deletion (IS1326 in IS1326::IS1353 is almost certainly functional) rightwards towards or past the IRt end of the integron while retaining the IS (Fig. Tn3.7Q and Fig. Tn3.7 W); to Tn2607 by insertion of Tn2601 (probably similar to Tn3) into the mer genes; to Tn2424 by insertion of IS161 to first generate Tn2425 and subsequent acquisition of two integron cassettes aacA1 and catB2 (Fig. Tn3.7Q and Fig. Tn3.7 X); and to Tn2603 by insertion of an oxaA1 cassette.
Tn1721 and (tandem) amplification of the tet genes
Tn1721 (Fig. Tn3.7 K) carries resistance to tetracycline (tet), is present on plasmid pRSD1 and is capable of undergoing amplification to generate tandem repeats [69]. It was isolated by transposition to a lambda phage followed by a further transposition event onto plasmid R388 [25] where it retained the ability to amplify [25] .
Amplification was identified using restriction enzyme mapping (Fig. Tn3.7 Y) which showed a duplication of an EcoRI fragment and presumably occurs via replication slippage or unequal crossing over during replication between the full tnpA gene and the 5’-end tnpA segment at the right end of Tn1721. Indeed, amplification was shown to depend on the host recA gene [70].
The Tn163 Clade
There are 39 members of this clade (May 2021). Two (TnSku1 [Tn7197] (CP002358.1) and TnAmu_p (NC_015188.1) have acquired toxin/antitoxin gene pairs and most members (Fig. Tn3.8 A; Fig. Tn3.8 B) encode divergent tnpR and tnpA genes. There are a number of members without passenger genes as in the Tn21 clade (e.g. Tn6137, TnMex22[ Tn7165], TnMex38 [Tn7166], TnChe1, [Tn7155], TnAmu1 [Tn7138 ], TnAli20 [Tn7136], Tn6122, Tn3434).
One small related group (Tn6137, Tn6136, Tn6134, Tn6138) all identified within the hexachlorocyclohexane-degrading bacterium Sphingobium japonicum UT26 genome [71] show evidence at the DNA sequence level of several recombination events including acquisition of an sdr passenger gene and exchange of tnpR and tnpA by exchange at a location at which res should occur (Fig. Tn3.8 C).
Alignment against Tn6136 (Fig. Tn3.8 Ci) shows that Tn6137 carries the left half while Tn6134 carries the right section while Tn6137 carries the right while Tn6134 carries the left segments of Tn6138 (excluding the passenger gene insertion). Although the res sites have yet to be defined in detail, comparisons clearly show sequence divergence in this region (Fig. Tn3.8 Cii). Both Tn6134 and Tn6138 carry the same passenger gene (Fig. Tn3.8 Ciii) whose insertion has occurred proximal to IRL (Fig. Tn3.8 Civ).
The ancestor of another group of related transposons, the Tn5393 group (Fig. Tn3.8 D), appears to be Tn5393c (AY342395.1; Pseudomonas syringae pv. syringae plasmid pPSR1) which underwent an insertion of Tn5501.6 to generate Tn5393.1 (MF487840.1; Pseudomonas aeruginosa PA34), of IS1133 to generate Tn5393 (M95402; Erwinia amylovora plasmid pEa34) (Fig. Tn3.8 E) and of a complex set of mobile elements to generate Tn5393.4 (AJ627643; Alcaligenes faecalis).
Tn5393 also gave rise to a number of other derivatives: Insertion of Tn3 into its transposase gene generated Tn5393.7 (LT827129; Escherichia coli strain K12 J53); insertion of Tn10 into IS1133 to generate Tn5393.2 (CP030921; Escherichia coli KL53 plasmid pKL53-M) (Fig. Tn3.8 F) followed by insertion of IS903 to generate Tn5393.11 (CP000602; Yersinia ruckeri YR71 plasmid pYR1); insertion of Tn10 in res to generate Tn5393.8 (CP002090; Salmonella enterica subsp. enterica plasmid pCS0010A).
There are also 4 examples carrying derivatives of Tn5 inserted into tnpA. They have an identical 3’ junction. In Tn5393.12 (KM409652; Escherichia coli REL5382 plasmid pB15), carries a complete Tn5. A second, Tn5393.13 (AB366441; Salmonella enterica subsp. enterica serovar Dublin plasmid pMAK2) is derived from Tn5393.12 by insertion of Tn2 into the IS1133 copy. In Tn5393.3 (LT985287; Escherichia coli strain RPC3 plasmid: RCS69_pI) the Tn5 insertion is a partial head-to-head Tn5 dimer, and in the other, Tn5393.10 (CP019905; Escherichia coli MDR_56 plasmid unnamed 6), insertion(s) and deletion(s) have occurred leaving only a partial Tn5 sequence.
Finally, Tn5393 also gave rise to Tn5393.9 (KU987453; Klebsiella pneumoniae 05K0261 plasmid F5111) by multiple insertion including a type II intron, IS5708, ISCR1, ISEc28, ISEc29 and Tn2. A number of intermediate structures have yet to be identified but can probably be found in the large number of Tn5393 derivatives in the public databases. This group of Tn163 clade members have undergone a large number of modifications and constitute a broad network of related elements.
The Tn4430 Clade
At present (May 2021) this clade is composed of only 11 examples (Fig. Tn3.9 A). One example, Tn4430 (X07651.1), encodes a tnpI gene and a res site with its associated organization but no passenger genes. The others encode a tnpR gene (Fig. Tn3.9 B). There are two small groups: Tn1546 which carry vancomycin resistance genes, and Tn6332 which carry mercury resistance genes.
The Tn1564 Vancomycin Resistance Group
Resistance to Vancomycin in Enterococci appeared in 1988 [72], was shown to be transmissible [73][74] and carried by a transposon, Tn1546 (M97297.1) [75]. The relationship within the Tn1546 vancomycin resistant transposons is relatively simple and the result of insertions/deletions mediated by several different insertion sequences: Tn1546.2 (AB247327) is derived from Tn1546 [75][76] by insertion of IS1216E between vanYA and vanXA and Tn1546.1_p (KR349520.1) appears to be derived from Tn1546.2 by insertion of IS1251 between vanHA and vanSA and a neighboring deletion to the right of IS1216E bringing vanYA and vanXA closer to each other. Other examples identified in surveys of vancomycin-resistant Enterococci from human and other animal sources also include insertions of ISEf1, IS1542 and IS19 [77], in addition to a number of other IS1216 insertions (often in multiple copies and accompanied by neighboring deletions) [76][78]. A number of these insertion/deletion derivatives have been identified from several sources and different geographical locations [76][77][78][79] (Fig. Tn3.9 C).
The Mercury Resistance Group
Within the mercury resistance group (Tn6294-LC015492.1, Tn5084-AB066362.1, Tn6332-LC155216.1 and TnMERI1-LC152290 – note that we have reconstituted the left end by comparison with Y08064; Fig. Tn3.9 D), the mercury resistance genes are expressed to the left while TnpR and TnpA are expressed to the right. All four carry additional copies of merB and merR. Huang et al [85] have shown that expression of the mercury resistance genes of TnMERI1 is driven by three promoters (Fig. Tn3.9 E). Comparison with Tn6294 suggests that the mercury gene set has been exchanged by recombination at the level of the res site (Fig. Tn3.9 D).
The sequences of two closely related members of the same group, Tn5083 and Tn5085, are incomplete [86].
The Tn3 Clade
This clade includes the classical Tn1, 2 and 3 (see Historical) as well as Tn1000. There are 29 examples of the Tn3 clade (of which 26 can be found in TnCentral) (Fig. Tn3.10 A) which fall into two subgroups. The majority have divergently expressed tnpR and tnpA and most carry passenger genes (Fig. Tn3.10 B). The res sites of each sub-group show significant similarity (Fig. Tn3.10 C). A number carry toxin-antitoxin genes (TA) generally located between the divergent tnpR and tnpA. These are of two types (Fig. Tn3.10 A) and appear to be specific for each subgroup. Passenger genes can be located upstream of downstream of the tnpR/tnpA transposition module (Fig. Tn3.10 B). All except two encode tnpR type resolvases. The two which do not, TnBth4 and Tn5401, also encode a TA module.
Importance of ISEcp1 in bla CTX-M-expression
There are examples of members of the Tn3 clade which carry insertions of ISEcp1-like sequences (see: IS1380 family) closely upstream of a bla-CTX-M gene. Indeed, upstream insertion of ISEcp1 derivatives have been identified associated with a number of different bla-CTX-M variants in both Tn3 and other groups [87][88][89][90][91][92]. In some examples, this is limited to an isolated right end [92] which is responsible for expression of the bla-CTX-M gene by providing a mobile promoter [93].
The Tn3 group
Tn3, Tn1, Tn1MER, Tn2, Tn2.1 and Tn3.1. all carry a probable internal IR upstream of the bla gene (Fig. Tn3.10 D) which acts as a hotspot for IS231A insertion and was initially observed in the bla gene of plasmid pBR322 [94]. Tn2 and Tn2.1 are identical except for the ISEcp1 insertion which also carries an internal IS1 insertion (Fig. Tn3.10 E). Note that an ISEcp1 promoter drives bla CTX-M-expression. There are a number of closely related derivatives (e.g. Tn6339-MF344565) in which the IS1 copy appears to have been involved in small rearrangements of the ISEcp1 copy while maintaining the ISEcp1 promoter. Three examples carry a number of integron cassettes without either the integrase gene, the Tn402 ends or the Tn402 transposition genes that are often associated with integrons in the Tn21 clade.
Inspection of the alignment (Fig. Tn3.10 E) shows that apart from insertion of different mobile elements, the major sequence variations occur in the region of the res sites, the 5’ ends of tnpA and tnpR as had been previously noted for Tn1, 2 and 3 [95] (for res, see Fig. Tn3.10 C) and an evolutionary pathway involving a combination of homologous and resolvase-mediated recombination has been proposed.
This can be detected by the distribution of SNPs on each side of the res site (e.g. Tn1331 and Tn1332). In this respect, the integron carrying Tn6238 is more similar to Tn3 while Tn1MER, Tn1331, and Tn1332 are more similar to Tn1 and Tn2.1 resembles Tn2.
The Xanthomonas group
This group except for TnPsy39 (Tn7187), all members of this group in the tree carry the same TA pair and the passenger genes are located to the right of the transposition module. The Xanthomonas transposon cluster (Fig. Tn3.10 F) are closely related and differ essentially by insertion of ISXac1 and ISXac5 (Fig. Tn3.10 G) as well as deletions (in particular of the res site in TnXc4.2 [Tn7212]). TnXc4.1 [Tn7211], although having an organisation identical to that of TnXc4 [Tn7210] has undergone significant sequence divergence along its entire length. TnThsp9 [Tn7202] also shows sequence variation within the region carrying transposition and TA functions (but includes mercury genes instead of plant pathogenicity functions while TnPsy39 [Tn7187] only exhibits similarity in the TnpA gene.
All members of the second cluster, which encode for the same TA gene pair as the Tn3 group (Fig. Tn3.33A), also carry mercury resistance genes although these have undergone some rearrangements and sequence divergence (Fig. Tn3.10 H) and are also divergent from those present in TnThsp9 (Tn7202).
The Tn3000 Clade
This clade is composed of nearly 30 members (25 in TnCentral) all of which encode TnpR resolvases and carry tnpR-related res sites. Most also encode TA gene pairs and these are of three types (Fig. Tn3.11 A).
The Tn5501 cluster.
There are a number of Tn5501 examples (Fig. Tn3.11 B). All have their passenger genes located upstream of the transposition module and all except TnPysy42 [Tn7188] and Tn5501.12 encode the same parE/parD TA genes (Fig. Tn3.11 A). Tn5501.12 appears to have acquired different TA genes (HTH_37, GP49) by recombination at the res site (Fig. Tn3.11 C).
The relationship between members of the cluster is shown in Fig. Tn3.11 C. Most have retained the same transposition and TA modules but vary in the type of passenger genes they carry. They all carry deletions with respect to Tn5051.3. For 8 of these, the right junctions of the deletions are close but not identical (Fig. Tn3.11 Di and Dii). All leave the TA module intact. In only one example, the toxin gene has undergone deletion leaving the antitoxin intact (Fig. Tn3.11 Diii). The left junction is less clear and difficult to interpret.
A number of Tn5501 derivatives are related by IS insertions and deletion (Fig. Tn3.)
Finally, a small group of Tns which, like Tn5501.12, all carry the HTH_37/GP49 TA pair is shown in Fig. Tn3.11 F. It appears that there has been an exchange between a Tn5501.5-like transposon and a derivative of Tn4662a (lacking the ISAs20 insertion) by recombination at the res site to generate Tn5501.12.
Clinical Importance of Tn4401
In the past decades, carbapenemase-producing Enterobacteriaceae (CPE) have appeared that are resistant to most or all clinically available antibiotics, including carbapenems, which are often considered the antibiotics of last resort [97]. The 10kb transposon, Tn4401 has been instrumental in the spread of the carbapenem resistance gene blaKPC. It was described in 2008 in a number of clinical isolates of Klebsiella pneumoniae and Pseudomonas aeruginosa from the United States, Colombia and Greece [98][99].
Members of this small group have divergently expressed tnpR and tnpA genes located towards the left end and blaKPC towards the right end downstream from tnpA (Fig. Tn3.11 B) flanked by two different insertion sequences, ISKpn6 and ISKpn7 (Fig. Tn3.11 G). The ISKpn7 insertion had occurred within an additional Tn4401 IR. It was further observed that there were two “isoforms” of Tn4401: Tn4401a and Tn4401b. Tn4401a, isolated in the United States and Greece carried a 100bp deletion upstream of the bla gene compared to Tn4401b from Colombia. The Tn4401 backbone appears to have undergone a number of recombination events. A third derivative, Tn4401c [100], was found to carry a deletion of about 200 bp upstream of bla while in a fourth, Tn4401d [101], the ISKpn7 copy along with flanking DNA has undergone deletion to leave a 3’ segment of blaKPC and a 5’ segment of tnpA and therefore would not be capable of autonomous transposition.
Furthermore, analysis of a number of clinical isolates from different regions of the United States which exhibited various levels of carbapenem resistance, revealed deletions of different extent in the region upstream of blaKPC [102]. Closer analysis using RACE (Rapid amplification of cDNA ends) to locate transcriptional start points revealed 3 (possibly 4) promoters, one of which had been generated from the -35 element located in the IR of the inserted ISKpn7 (as is characteristic for a member of the IS21 family (see IS21 chapter; formation of hybrid promoters figure IS21.1).
The Tn4651 Clade
The Tn4651 mix of radically different structures
This Tn3 family clade (Fig. Tn3.12 A) contains members with very diverse structures (Fig. Tn3.12 B). They fall into three major clusters. Two encode the tnpT/S/rst while the third encodes the tnpR/res system.
The tnpT/S/rst clusters
In the first tnpT/S/rst cluster, mostly from the plant pathogen Xanthomonas (Fig. Tn3.12 C), TnXax1.1 [Tn7207] appears to have undergone res-recombination in which the upstream passenger genes and tnpT have been exchanged. TnpT is significantly different from the other four. TnXax1.3 [Tn7209] differs from the others (TnXax1 [Tn7206]; TnXax1.2 [Tn7208]; TnXax1.3 [Tn7209] in the 3’ region of tnpA and there is some variation in tnpS and tnpT.
TnXax1 derivatives [26] are generally vehicles for pathogenicity genes such as Transcriptional Activator Like Effectors (TALE genes), lytic transglycosilases (mtlB2) and genes (xop) involved in type III secretion system (TTSS) translocation of effector proteins into host plant cells [107] (Fig. Tn3.12 C). TnXax1 derivatives can include IR which are significantly longer (72/92 bp) than the 38-40bp characteristic of the Tn3 family (Fig. Tn3.12 D) although the functional significance of this has not been investigated. The IR also terminate in a GAGGG pentanucleotide. The left end of group members is quite variable (Fig. Tn3.12 E) while their right ends appear more homogeneous (Fig. Tn3.12 F).
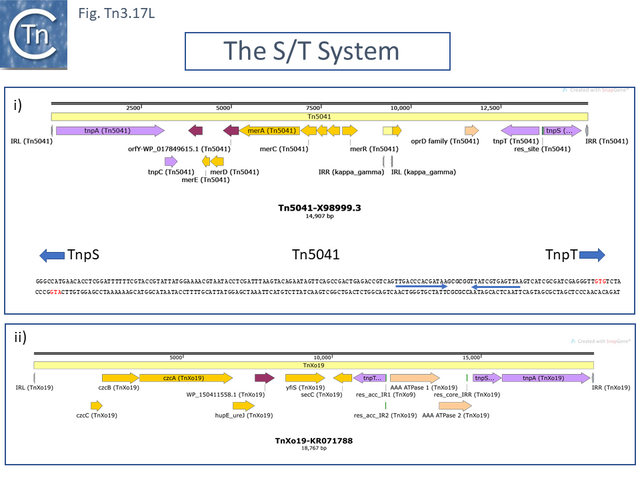
The second tnpT/S/rst cluster is characterized by Tn4651, a toluene-catabolic transposon identified in from Pseudomonas putida plasmid pWW0 [108]. In addition to the tnpS/T resolution system, it encodes an additional small transposition-related gene, tnpC which impacts cointegrate formation.
Using, Tn4652, a Tn4651 deletion derivative lacking the toluene-catabolic genes [108], TnpC was shown to regulate TnpA expression post-transcriptionally [109]. Moreover, the host protein IHF binds to sites in both Tn4652 ends (Fig. Tn3.12 G) [110][111]. These overlap the region protected by TnpA binding [111] and binding positively regulates both tnpA transcription and TnpA binding to the terminal IRs. Indeed, transposase binding to the IRs in vitro was shown to occur only after binding of IHF [111]. TnpA protects an extensive region encompassing the IRs and 8-9 bp of flanking DNA (Fig. Tn3.12 G). Tn4652 transposition appears to be elevated in stationary phase, involves the stationary phase sigma factor, sigma S [112], and is limited by the levels of IHF [111] whose level is increased in stationary phase. Another DNA chaperone host factor, FIS, has a negative effect on transposition, apparently by competing for IHF binding [111][113].
IHF and FIS have been implicated in other transposition systems such as IS10 (see: IS4 family). Moreover, Tn1000 (Tn3 clade) carries an IHF binding site proximal to each IR which acts copoperatively to increase TnpA binding and immunity [114][115]. One additional interface with host physiology is the observation that the CorR/CorS two component system regulates transposition positively [116].
Other members of the cluster include: Pseudomonas sp. mercury resistance transposon Tn5041 [117][118] ; Tn4676, a long (72,752bp) and complex Pseudomonas resinovorans carbazole-catabolic transposon from plasmid pCAR1 [119][120]; and Tn4661, a Pseudomonas aeruginosa cryptic transposon [28]. All include tnpA, tnpC and the tnpS/T resolution system.
Tn5041 transposition has also been addressed experimentally[117][121] and was observed to be host-dependent [121]: it occurred in the original Pseudomonas sp. KHP41 host but not in P. aeruginosa PAO-R or in Escherichia coli K12. Interestingly, transposition in these strains was found to be complemented by the Tn4651 transposase gene (tnpA) and the region which determines this host dependence was mapped to a 5’ tnpA gene segment by construction of hybrid Tn5041-Tn4651 tnpA genes. Tn5041 apparently acquired its mer operon from a derivative of Tn21 or Tn501 [121]. It is reported to be preceded by a 24 bp element with 75% sequence similarity to the outermost part of IRs typical for Tn21-like transposons.
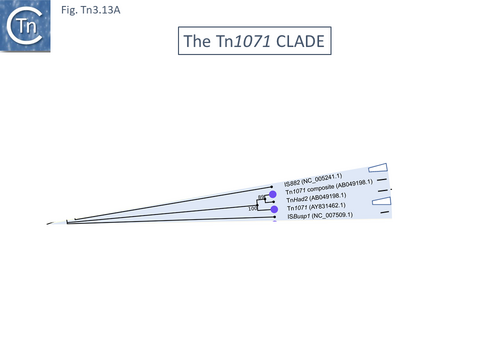
The Tn1071 Clade
The Tn1071 group.
Members of this small group are often associated with xenobiotic catabolism and other “exotic” functions (Fig. Tn3.13 A).
Tn1071 itself (Fig. Tn3.13 Bi), the founding member, was identified as part of a compound transposon, Tn5271, in Comamonas testosteroni where it flanks a chlorobenzoate catabolic operon in [123]. It is unusual since it carries only tnpA and not tnpR, has unusually long (110bp) IR (Fig. Tn3.13 Bii) and was first described as IS1071. Two other members of this small group, IS882 from Ralstonia eutropha H16 megaplasmid pHG1 encoding key enzymes for H2-based lithoautotrophy and anaerobiosis [124] and ISBusp1 (aka ISBmu13; NC_007509.1) from the Burkholderia multivorans ATCC 17616 genome [125], were also originally identified as IS. Their structure fits the definition of an IS since they all contain a single transposase open reading frame located between two IR.
A limited functional analysis of Tn1071 transposition is available [126]. It was only able to transpose at high frequencies in two environmental β-proteobacteria Comamonas testosteroni and Delftia acidovorans but not in Agrobacterium tumefaciens (α-proteobacteria) or Escherichia coli, Pseudomonas alcaligenes and Pseudomonas putida (all γ-proteobacteria). These studies showed that Tn1071 generates cointegrates as a final transposition product since it has no resolution functions, produces 5bp DR on insertion and requires the entire 110bp IRs for activity. This is therefore in contrast to many other Tn3 family members which only require the 38 bp IR.
The absence of a resolution system implies that, like IS26(see: IS6 family) , Tn1071 probably forms “pseudo-compound transposons” [127][128][129]. In these structures the flanking Tn1071 copies must be in direct orientation as a consequence of the homologous recombination event required to resolve the cointegrate structure. Transposition is initiated by one of the flanking IS to generate a cointegrate structure with three Tn1071 copies (similar to those generated by the IS6 family of insertion sequences; Fig. IS6.8B). “Resolution” resulting in transfer of the transposon passenger gene requires recombination between the “new” IS copy and the copy which was not involved in generating the cointegrate.
The implications of this model as for IS6 family members are that the transposon passenger gene(s) are simply transferred from donor to target molecules in the “resolution” event and are therefore lost from the donor “transposon” leaving a single Tn1071 copy in the donor plasmid. However, it is possible that both Tn1071 copies are used in transposition in which case the cointegrated would be expected to contain two directly repeated copies of the entire transposon sat the donor/target junctions.

A significant number of Tn1071-associated xenobiotic-degrading genes on many catabolic plasmids have been documented by population-based PCR [130][131][132] and genetic studies [133][134].
Tn5271 itself is widely distributed in bacteria isolated from a large ground water bioremediation site [132] and plasmid derivatives carrying the transposon together with a third Tn1071 copy in an inverted orientation were also identified. The interstitial DNA segment between the old and new copy in these derivatives was also inverted as expected from intra-molecular transposition events [132] (Fig. for intramol transposition).
A number of additional potential compound transposons have been identified although these may be inactive: a >28kb transposon, Tn5330 (AF029344), from Delftia acidicorans [135] carries the entire 2,4-dichlorophenoxyacetic acid degradation pathway and, although the sequence data for the flanking Tn1071 copies is not complete, both carry inactivating insertions of IS1471; a similar ~48 kb transposon (NC_005793) with 5bp flanking DR from Achromobacter xylosoxidans plasmid, pEST4011, also carries identical IS1471 inactivating insertions in each flanking Tn1071 copy [136] and a 7kb internal tandem duplication compared to the Delftia acidovorans transposon.
When analyzed in more detail, these genes are sometimes flanked by Tn1071 copies in direct repeat as in the original Tn5271 but are found in more complex Tn1071-based structures.
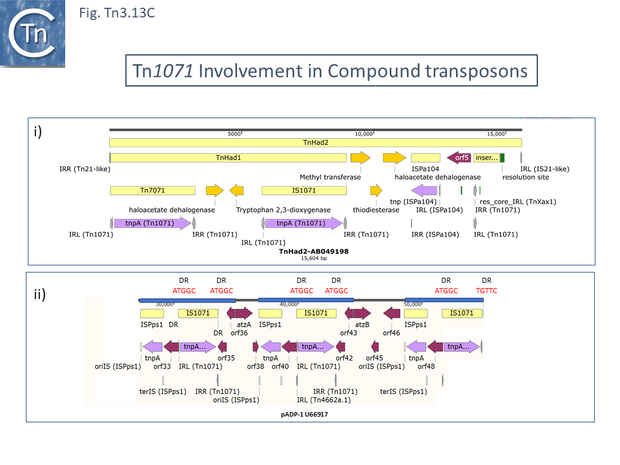
TnHad2 [137] (Fig. Tn3.13 Ci), for example, from a Delftia acidovorans haloacetate-catabolic plasmid, pUO1, carries a nested copy of a potential Tn1071-based compound transposon, TnHad1 which does not carry flanking DR. TnHad1 is inserted into a larger structure, TnHad2 with flanking 5bp DR, typical Tn3 family ends related to those of Tn21 but no apparent dedicated transposase except that of the Tn1071 copies.
The authors state that TnHad2 was unable to transpose as judged by a “mating out” assay using the plasmid R388 as a target. However, The TnHad2 Tn21-like IRs were found to be active in transposition if supplied with Tn21 but not with Tn1722 transposition functions [137]. TnHad2 also appeared to carry a functional res site.
Tn1071 also flanks atrazine degrading genes in plasmid Pseudomonas pADP-1 (U66917) [138] in a structure with three directly repeated Tn1071 copies intercalated with three copies of an IS91 family member, ISPps1. These are apparently generated by duplication events since regions with identical sequence stretch from the oriIS end of ISPps1 through Tn1071 and terminate just before the atz genes (Fig. Tn3.13 Cii). The repeated regions also includes the DR sequences at each Tn1071 except for that at the far right.
There are a number examples of other structures with multiple Tn1071 copies and in a large proportion of these cases, the multiple copies occur in direct repeat. They are associated with plasmids which degrade the phenylurea herbicide linuron e.g. pBPS33-2 (CP044551) [140] and have been isolated from a variety of bacteria with the capacity to degrade a wide range of chlorinated aromatics and pesticides [130] or p-toluene sulfonate (PTSA) where they flank the PTSA genes in plasmid pTSA (AH010657) [141].
MITES, MICs and TALES
Many TE families also include non-autonomous transposable derivatives with no transposition related genes. These are simple and composed of two correctly oriented ends with or without an intervening passenger gene and are called MICs (Minimal Insertion Cassette) and MITEs (Miniature inverted-repeat transposable elements) respectively. For Tn3, related MITEs are known as TIMEs (Tn3-Derived Inverted-Repeat Miniature Elements) [142][143].
Studies have shown that Xanthomonas genomes are often havens for MICs carrying genes involved in pathogenicity towards their host plants [26]. A number of Tn3 family structures were identified in a conjugative plasmid, pXac64 (CP024030), of the principal pathogen of citrus trees, Xanthomonas citri, an important economic problem (e.g., reference [144]) (Fig. Tn3.14 A). The plasmid includes two Tn3 family transposons, TnXc4 (Tn7210) and TnXac1.4 (Tn7206) and a MIC (MIC XAC64.T1; 3948bp) which carries a TAL effector gene. Other TAL effector-carrying MICs can be identified in other Xanthomonas plasmids such as pXac33 (CP008996) [145] (two TAL-carrying MIC: MIC XAC33.T1, 3739bp, and MIC XAC33.T2, 3538bp; and the Tn3 family transposon TnXc5) and from the Xanthomonas fuscans plasmid pplc XAF (FO681497) [146] (a single MIC, MIC XAF.T1 ,3768bp, and a 10kb MIC with a number of virulence genes. Some MICs, e.g. MIC XAC33.T1 (Fig. Tn3.14 B right), are flanked by 5bp DR, a hallmark of Tn3 family transposition.
A global analysis of TAL effector genes in (Fig. Tn3.14 C) within the Xanthomonas genus (available in 2014) identified a large number which were flanked by Tn3-like IR although a some carried a single identifiable IR while others failed to exhibit clear IRs [26].
Inspection showed that the chromosome of Xanthomonas citri strains do not carry identifiable TAL-carrying MICs but those of X. oryzae carry relatively high numbers [26]. A smaller number of MICs carrying other pathogenicity-related genes are also observed (e.g. Type III Xop genes) It is notable that the majority of the TAL-associated MICs occur as two or more tandem copies. These are listed for three example genomes, X. oryzae PXO99A, MAFF and KACC in Fig. Tn3.14 D.1-3.
Those where no IRs could be detected at either end are shown simply as open reading frames. In each case, the DNA segment between tandemly repeated MICs is identical (Fig. Tn3.14 E), suggesting that the tandem dimers and multimers arose by amplification possibly via replication slippage and unequal crossing over [26] (Fig. Tn3.14 F). Another characteristic is that they are often flanked by transposase genes raising the possibility that their appearance at different chromosome locations (“radiation”) has occurred by transposition of a single ancestral MIC. This might have been mediated either by flanking transposable elements or by complementation from a Tn3 family transposase. In many cases, one of the terminal MICs is truncated and does not exhibit an IR and could often be attributed to insertion of an IS.
It is clear that this “radiation” of TAL-associated MICs does not only occur by transposition. In one case (Fig. Tn3.14 Ei, Eii and Fig. Tn3.14 G) an entire DNA segment containing a tandem MIC dimer (MIC P.T11-MIC P.T13) appears to have been translocated together with surrounding genomic sequences with MIC P.T13 undergoing deletion to generate MIC P.T11-MIC P.T12.
This variability in MIC sequence can be observed within the longer arrays (e.g. Fig. Tn3.14 E viii) suggesting that diversification follows amplification. This is due to changes in the TAL genes. TAL proteins are composed of conserved N-terminaland C-terminal regions separated by a variable number of 34 amino acid repeats (Fig. Tn3.14H) which can number between 1.5 and 35.5 tandem copies. Each repeat includes a pair of adjacent amino acids capable of recognizing a single base in a DNA sequence (Fig. Tn3.14 H; Fig. Tn3.14 I) e.g. [148][149][150]. A tandem array of repeats therefore enables the TAL protein to recognize specific sequences within the target plant genome. This is illustrated by the TAL effector carried by MIC P.T14 (Fig. Tn3.14 J) which includes 19.5 such repeats. The TAL effectors encoded by other members of this cluster (Fig. Tn3.14 Eviii), MIC P.T15, MIC P.T16, MIC P.T17, MIC P.T18, which have presumably all arisen by amplification of a single ancestral MIC, each carry a different number of repeats and vary in their sequence recognition properties.
It is interesting to note that while the amino acid repeats are always maintained in phase, certain TAL effectors have undergone removal of a single amino acid while another has acquired a short insertion. These changes might be expected to influence the capacity of the proteins to recognise their cognitive DNA sequence.
Diversification can also be observed between clusters in related X. oryzae strains such as MAFF and KACC.
Strain PXO99A and MAFF share the cluster MIC P.T15, MIC P.T16, MIC P.T17 (Fig. Tn3.14 Ki; Fig. Tn3.14 L). Both clusters have identical genomic environments (with some sequence variation) and the inter MIC sequences are identical. Not only has there been a large deletion of MIC P.T18 in the MAFF cluster, but sequence variations are apparent along the entire cluster length both within and between the clusters potentially modifying the DNA sequence recognition properties. Strain MAFF and KACC also share a cluster (MIC M.T2 and MIC M.T3).
Further analyses and experimental approaches are necessary to fully understand the role of MICs in the dispersal and diversification of these important instruments of Xanthomonad virulence, the TAL effectors.
Acquisition of Passenger Genes.
Tn3-family transposons carry large and diverse and diverse sets of passenger genes (e.g. Fig. Tn3.3). These have been acquired by a number of different processes.
Tn402 and integron platforms.
One major source of antibiotic passenger genes has been by ancestral insertions of Tn402 derivatives which have often “decayed” to lose their transposition properties but have retained their abilities to acquire (and lose) integron gene cassettes (Fig. Tn3.7 G; Fig. Tn3.7 I; Fig. Tn3.7 J; Fig. Tn3.7 R; Fig. Tn3.7 S; Fig. Tn3.7 U; Fig. Tn3.7 V; Fig. Tn3.18C).
Additional TE
A second pathway to acquisition is by insertion of additional transposable elements with, or without rearrangement (Fig. Tn3.7 U; Fig. Tn3.8 E; Fig. Tn3.8 F). It is also interesting to note that there are a number of cases in which additional IR appear within certain structures (e.g. Fig. Tn3.10 D; Fig. Tn3.11 G) such as Tn3 [94] and Tn501 [155] raising the possibility that these have been involved in generating the host transposon.
Recombination at res.
A third major pathway to passenger gene acquisition is by inter-transposon exchange via res sites (see: Resolution). This was first suggested to explain the formation of Tn501, by exchange of a transposition module with a Tn1721-related transposon [155]. It was later observed by Kholodii and coworkers [156][157] and called “shuffling”, by Yano et al., [158] and by others [34].
As judged by the analyses included here, this seems to be a recurring type of event and can be found in members of most clades (Tn21: Fig. Tn3.7 H; Fig. Tn3.7 M; Fig. Tn3.7 N; Tn163: Fig. Tn3.8 Ci and Fig. Tn3.8 Cii; Tn4330: Fig. Tn3.9 D; Tn3000: Fig. Tn3.11 C; Fig. Tn3.11 F; and Tn4561: Fig. Tn3.12 C). This type of behavior can also lead to “suicide” of a transposon in which the transposition module is removed by res recombination with a site outside the transposon [159].
Mercury Resistance: a Major Passenger Gene Group
The Mercury Operon and the Tn3 family
Not surprisingly, bacteria carrying mer operons are particularly abundant in areas with increased mercury concentrations such as mercury mines and contaminated soil or water [160][161][162] and it was suggested that mercury resistance is an ancient system as reflected by a wide geographical, environment and species range and that it evolved as a response to increased levels of mercury in natural environments resulting, for example, from volcanic activity [163].
It is certainly present in the Murray collection [164], a collection of Enterobacteriaceae isolated in the pre-antibiotic era, as part of transposons Tn5073 and Tn5074 which show high similarity to present day examples such as Tn5036 and Tn1696 (Tn3 family members of the Tn21 clade) and Tn5053 (a Tn402 family member of the Tn5053 clade) and Tn5075 respectively [62].
Although Tn3 family members carry a large variety of passenger genes, mercury resistance is found repeatedly within the family and is thought to be one of the first sets of passenger genes to be acquired (Fig. Tn3.6) and appears in precursors of the major groups of antibiotic resistance carrying Tn3 family members (Fig. Tn3.7 G). Mercury resistance operons were proposed to have been acquired at least twice [43](Fig. Tn3.6): once by an ancestor of Tn21 and once by an ancestor of Tn501. Their acquisition presumably predates the acquisition of antibiotic resistance integron platforms since a number of mercury resistance Tn3 family transposons have been identified and, in at least two cases, Tn21 and Tn1696 (whose mer genes appear to fall largely into different groups; Fig. Tn3Bi-vii), clear precursors devoid of integrons (Tn5060 [45] and Tn20 and Tn1696.1 respectively) have been identified. Mercury resistance genes are found in a number of Tn3 family clades (Fig. Tn3.15 B and Fig. Tn3.15 Bi-vii). These include Tn3, Tn21, Tn163, Tn4430 and Tn4651. Those associated with the Tn21 clade occur upstream of, and are generally expressed towards, tnpR (Fig. Tn3.7 G); those of the Tn3 clade are located downstream of tnpA (Fig. Tn3.15 C) and in those carrying the tnpS/T genes, they are between the transposase module and the tnpS/tnpT module (Fig. Tn3.15 D).
A survey of 29 functional mercury resistance transposons isolated from Gram negative bacteria in environmental isolates revealed that the most widespread of transposons belong to two types: transposons of the Tn21 clade of the Tn3 family and relatives of Tn5053, a member of the Tn402 family [157][165]. In addition, Yurieva et al [157] identified a third group, related to Tn5041, a member of the Tn4651 clade They also identify “mosaic” mer operonswhich, they suggest, are generated by homologous recombination between short DNA sequences. While MerR appears to be very similar between different mer operons, while MerA showed a higher degree of mosaicism as did MerT and MerP to some extent [157].
The Mercury Operon: Organization, Regulation, and Resistance Mechanism
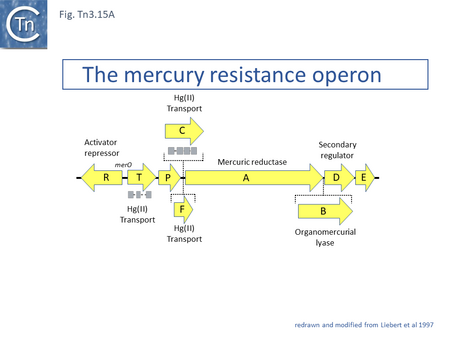
The mechanism underlying mercury resistance has been extensively reviewed a number of times [58] . Briefly, mercury resistance in gram-negative bacteria results in the release of gaseous mercury Hg0. Mercury salts (HgII) are captured by the periplasmic MerP, transferred across the periplasm to the inner membrane proteins MerC or MerT and then across the cytoplasmic membrane to the mercuric reductase, MerA which converts it to the volatile Hg0. The operon is regulated by two genes, merR and merD (Fig. Tn3.15 A). The order of these genes is generally merT, merP, merC, merA, merD and merE. merR is located upstream and is transcribed in the opposite direction with overlapping promoters. Binding of MerR represses expression of the operon and of itself. Interaction with Hg(II) releases MerR repression of the mer structural genes permitting their expression without significantly impacting on its autorepression [166] and its interaction with RNA polymerase creates a pre-transcription initiation complex [167].
The product of the secondary regulator gene, merD [38], appears to play a role in down-regulating the mer operon [168]. It binds weakly but specifically to the merOP region and DNase I footprinting identified a common operator binding sequence for both MerR and MerD [168].
The genes essential for mercury resistance were identified as merR, merT, merP and merA [169]. An additional mercury ion transmembrane transporter gene, merE (UniProtKB - D4N5J4) involved in the accumulation of methyl-mercury [58][170] is often present. Not all mercury operons include merC and some have a gene, merF [171], an alternative mercury ion transmembrane transporter (UniProtKB - Q1H9Y3). Some also include a mercury lyase gene, merB, involved in resistance to organo-mercury [172][173].
The Mercury Operon: Diversity in various Tn3 family clades.
The mer carrying Tn3 family members (Fig. Tn3.15 B) all lack merF. Most examples carry a full mer gene complement although a small group (Tn501, Tn511, Tn1412, Tn4378 and Tn4380) lack the merC gene and only 3 (Tn5084, Tn6294, Tn6332), all members of the Tn4430 clade) carry a merB gene and have a duplicated or partially duplicated mer operon.
Phylogenetic trees generated for MerR (Fig. Tn3.15 Ci and Cii), MerT (Fig. Tn3.15 Ciii), MerP (Fig. Tn3.15 Civ), MerA (Fig. Tn3.15 Cvi), MerD (Fig. Tn3.15 Cvii) and MerE (Fig. Tn3.15 Cviii) reveal that, in general, Tn501-related mer genes group separately from those of Tn21 relatives. This provides some support for the hypothesis that the mer operon had been acquired at least twice. These groups are separated by mer genes from Tn402 family relatives.
Within the Tn21 clade, all members carry the mer operon upstream of tnpR with the direction of transcription to the right (Fig. Tn3.15 D top). merR, on the other hand, is transcribed in the opposite direction and terminates with a TAG codon within the IRL sequence (Fig. Tn3.15 D bottom) with one exception, Tn6023. On the other hand, for the few members of the Tn3 clade, the mer genes are located downstream of tnpA and are transcribed to the left (Fig. Tn3.15 E top) except for merR which is transcribed towards and terminates some distance from IRR (Fig. Tn3.15 E bottom), while for the unique Tn4651 member, the mer operons is located between tnpA and tnpS/T (Fig. Tn3.15 F).
The Mercury Operon: Tn21 in mer acquisition by Tn402?
It is worth noting that members of the Tn402 family Tn5053 mercury resistance subgroup carry a single copy of a sequence closely related to Tn21 IRL (Fig. Tn3.15 G top) located in a similar position with respect to the mercury operon as the resident IRL in the Tn21 group (Fig. Tn3.15 D and Fig. Tn3.15 G top and middle) (see [171]). There is some variability in the 10 C-terminal amino acid tail of the neighboring MerR protein (Fig. Tn3.15 G bottom) although the major part of MerR amino acid sequence is highly conserved. This raises the possibility that the mercury resistance genes carried by the Tn402 family elements was derived from an ancestral Tn21 group transposon.
Transposition Mechanism Overview
Early Studies
In early studies of Tn3 (Tn1 and 2) [12][13][14][15], Tn4651 [108] and Tn4430 [31][174] it was clearly demonstrated that Tn3 family transposition occurs in a two-step process involving a replicative step in which the transposon first couples the donor and target replicons by single strand transfer to create a forked allowing replication to generate a fully double stranded cointegrate structure followed by a site-specific recombination step, resolution, catalyzed by a dedicated enzyme, the resolvase (Fig. Tn3.2). While the resolution step for a number of Tn3 family members has been studied in exquisite detail (see Resolution below), study of the initial strand transfer and replication steps have proved problematic.
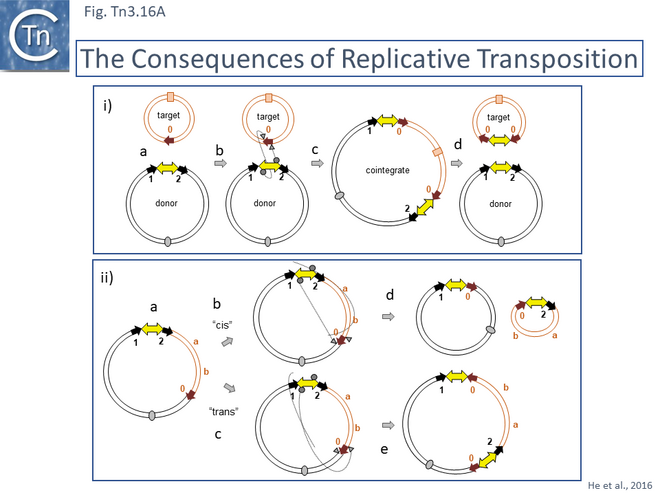
The consequences of these pathways are shown in greater detail in Fig. Tn3.16A. This underlines why not all Tn3 family transposition events yield transposons flanked by 5bp direct repeats. Figure Tn3.16Ai shows intermolecular transposition generating a cointegrate which, following resolution yields donor and target each with a single copy of the transposon in flanked by two DR copies. In intramolecular transposition, one pathway leads to a deletion while the other to an inversion. In neither case is the transposon flanked by direct target repeats. Tn3-mediated inversions and deletions of this type have been described a number of times with Tn3, Tn1, Tn2660 and Tn1721 [6][176][177][178][179][180][181].
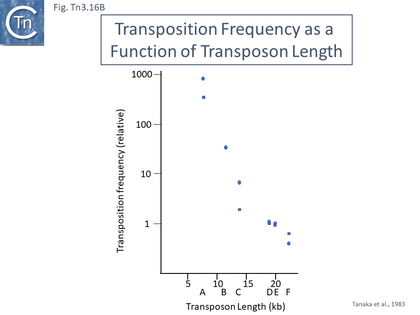
Early studies also demonstrated that, like a number of transposons, the transposition frequency of Tn3 family transposons appears to decrease exponentially with increasing length [41] (Fig. Tn3.16B). Tanaka and colleagues investigated Tn2603 and various derivatives ranging in length from approximately 5kb to 22.5kb from a number of different donor plasmids to both R386 and R388 target plasmids and noted a steep exponential reduction in transposition frequencies of over 1000-fold with increasing length. This observation would be more robust if transposition frequencies had been measured from the same donor plasmid and the transposons all had identical genetic contexts.
Replicative transposition
One of the major problems in studying transposition of Tn3 members is that their transposases, TnpA, are long (~1000 amino acids) and difficult to solubilize.
Interaction of transposase and transposon ends
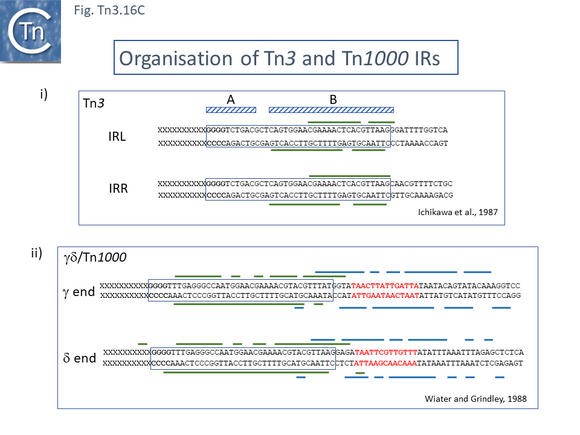
The Tn3 transposase, TnpATn3, was first purified in 1981 and shown to bind DNA in a salt resistant way [182] and one of the first attempts to investigate TnpATn3 activity in vitro [183] concluded that addition of ATP was necessary to obtain TnpATn3 binding to the Tn3 ends. However, in a subsequent article this was shown to be erroneous and probably due to a pH effect of the added ATP solution [184]. Purified TnpATn3 was observed to bind specifically to both IRTn3 and protect a sub-terminal DNA region within the IR (Fig. Tn3.16 Ci) in a heparin resistant manner a measure of its strong and highly sequence-specific DNA binding activity while another study using a different TnpATn3 purification scheme and DNA binding conditions [185] showed a much less sequence-specific protection which included the entire IRTn3 and a significant region of flanking DNA. Further functional analysis of the Tn3 ends [186] demonstrated that mutations in the first 10 IRTn3 base pairs (domain A) did not influence TnpATn3 binding while mutations in the 13-38 base pair region (domain B) inhibited binding (Fig. Tn3.16 Ci), behavior confirmed in a second study [187].
This is a similar functional architecture to the ends of other transposable elements (see: General Information/IS Organization/Terminal Inverted Repeats). In addition, the effects of mutations in the Tn3 ends on transposition in vivo [188] indicated that mutations in the TnpATn3 binding site have a stronger effect when present at both transposon ends than when located at only one end.
Similar binding studies have been undertaken for Tn1000 (γΔ) (Fig. Tn3.16 Cii). Protection against DNAse is more extensive than for Tn3 although this depends critically on the binding and digestion conditions [115]. The protection pattern is broadly similar with the tip of the terminal IRTn1000 remaining unprotected and protection extended to the inner end of the IR. Some weak protection occurred on the DNA region flanking the IR tip. In addition, however, the Tn1000 ends include a binding site for the host DNA architectural protein, IHF, and both proteins were found to bind cooperatively [115]. However, IHF appeared to downregulate Tn1000 transposition [114]. The juxtaposition of IHF sites and transposon ends has been observed in several other TE (see [189][190][191][192]).
Binding studies have also been carried out with the transposase of Tn4430, TnpATn4430, a Tn3 derivative which encodes a TnpI resolvase [193]. Here, it was necessary to use a mutant transposase (Fig. Tn3.16 Ei) which had been selected for a reduction in its transposition immunity (see Transposition immunity below) and which concomitantly showed an increase in transposition activity. Similar protection patterns (Fig. Tn3.16 Eii) were observed as with TnpATn3 and TnpATn1000: transposase binding protects the distal IRTn44300 internal region. The IR was divided into three regions (A, B1 and B2) based on sequence conservation, which largely correspond to the A and B regions of IRTn3 (Fig. Tn3.16 Ci).
TnpA functional domains
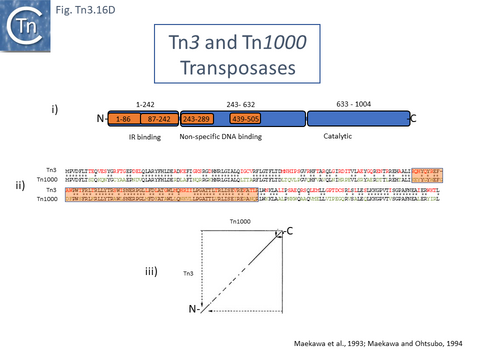
The TnpATn3 is 1004 amino acid residues long. Like many other transposases, it carries a DDE catalytic motif (General Information/Reaction mechanisms). Characterization of a series of fusions of TnpATn3 segments to β-galactosidase [194][195] (Fig. Tn3.16 Di) revealed that the N-terminal segment (residues 1-242) exhibited sequence-specific binding to the 38 base pair IR and that this region could be dissected into two sub-regions, amino acids 1-86 and 87-242, which showed non-specific DNA binding activity, implying that both were involved in sequence-specific end binding. The large central region also included two regions with non-specific DNA binding properties while the C-terminal region encodes the DDE catalytic site.
The region of TnpA involved in DNA sequence recognition for binding to the transposon IRs was further investigated using a series of hybrid TnpA genes carrying the N-terminal IR-binding region constructed between TnpATn3 and TnpATn1000 [195]. TnpATn3 and TnpATn1000 were found to share over 64% identity (Fig. Tn3.16 Dii). This enabled the definition of a region of TnpA which permits distinction between binding to an IRTn3 and an IRTn1000 [195] (Fig. Tn3.16 Dii). A dotplot comparison of tnpATn3 and tnpATn1000 nucleotide sequences indicated that the 3’ ends of both genes were conserved whereas the 5’ ends showed some variation (Fig. Tn3.16 Diii) [195].
A functional map of the Tn4330 transposase, TnpATn4430, was obtained by partial proteolysis with trypsin and chymotrypsin (Fig. Tn3.16 Di) [196]. This treatment indicated that, like TnpATn3, TnpATn4430 has three major domains: an N-terminal domain (amino acids 1-152) similar to a CENP-B DNA binding domain [197]; a central region (amino acids 153-682); and a C-terminal domain (amino acids 683-980) with an RNase H fold-like domain including the catalytic DDE triad. Like other members of the family, the distance between the second D and E residues is somewhat longer than in typical DDE transposases and has been called an insertion domain and is likely composed of alpha-helical structures [198]. The presence of insertion domains between the D and E residues observed in other transposases does not disturb the catalytic RNAse fold [198] and, in both cases studied in detail [199][200], performs crucial functions in the transposition chemistry specific for each element.
Cleavage and Strand transfer.
In spite of the extensive DNA binding studies, the biochemistry of Tn3 family transposition has proved refractory to detailed analysis. A single study with Tn3 [201] in vitro used a cell extract with high TnpA levels, a donor minimal plasmid replicon containing a mini transposon with Tn3 ends and a target molecule composed of concatemeric phage lambda DNA. Following the reaction, the phage DNA was packaged in an in vitro system and used to infect suitable recipient cells. The process yielded cells which appeared to carry large plasmids consistent with the formation of cointegrates. However, these were not physically characterized and the approach does not seem to have been developed further. Additionally, sequence-specific 3’ cleavage at the ends of a plasmid carried mini Tn3 derivative was observed with a cell-free extract containing TnpATn3 in a reaction which required Mg2+ and was stimulated by a host factor determined to be acyl carrier protein (ACP) [202]. A similar observation had been made for the Tn7 transposition reaction [203].
In a more recent a study using the mutant TnpATn4430 [193] an in vitro system including both strand cleavage and strand transfer was developed. The mutant TnpATn4430 carried 3 mutations (Fig. Tn3.16 Ei) selected for a reduced level of transposition immunity [196] but exhibiting a hyper transposition efficiency [193]. It was shown, using a gel shift assay and differentially fluorescently labeled IR, that this TnpA derivative formed two types of complex which appeared to be single end and paired end (SEC and PEC) species containing one or two IRTn4430 molecules bridged by the transposase.
Footprinting both types of complex revealed an identical pattern of DNase protection (Fig. Tn3.16 Eii) except for some additional weak protection of flanking DNA in the PEC. When probed with the 1,10-phenanthrolinecopper [(OP)2-Cu+] nuclease, the PEC showed significantly enhanced cleavage at the IR tip and in the DNA flank, particularly on the lower strand indicating a change of DNA conformation (Fig. Tn3.16 Eii).
Correct single strand cleavage at the 3’ end of the IR tip was observed in typical cleavage conditions as well as some double strand cleavage (3’ and 5’). This was examined using both wildtype TnpATn4430 and mutant derivatives with different transposition activities. The unexpected 5’ cleavage increase with increasing TnpATn4430 activity and when Mn2+ was used instead of Mg2+ indicating that this is an aberrant activity.
Furthermore, precleaved IR substrates were able to form a more stable PEC as observed in other in vitro transposition systems such as those of transposon Tn10 and bacteriophage Mu. The system was also shown to support strand transfer of a precleaved IR into a supercoiled target plasmid. Integration of both single and to a lower extent concerted integration of two IR was observed (Fig. Tn3.16 Eiii). Initial data have also suggested that TnpATn4430 binds preferential to DNA structures which resemble replication forks in vitro (cited in [24]) [207] and insertion appears to be influenced by replication of the target molecule in vivo (cited in [24]).
Some initial evidence was also presented suggesting that the PEC was composed of a pair of IRs and a single TnpATn4430 molecule. This has proved to be a misinterpretation of the data. In all other transposition systems, PEC complexes include two (or more) transposase molecules (e.g.[199][208][209]). Recent data both from Atomic Force Microscopy (AFM) and Cryoelectron microscopy demonstrates that the TnpATn4430 is indeed a dimer (B. Hallet personal communication; [210][211]).
Mechanism in the Light of Structure
A 3.6 Å average resolution cryoelectron microscopy structure has demonstrated that TnpATn4430 is indeed dimeric and has provided some insight into how it might function in transposition [210]. Moreover, using the hyperactive immunity deficient TnpA mutant it was possible to resolve a structure for the PEC which was composed of the transposase dimer and two double strand Tn4330 ends.
The structural model permitted a refinement of the TnpATn4430 functional modules obtained from partial proteolysis and footprinting (Fig. Tn3.16 Ei and Eii). Four DNA binding domains were identified (DBD1-4; Fig. Tn3.16 F top). DBD1,2 and 4 bind the IR in a sequence-specific manner. The first (N-terminal proximal) DBD1 establishes both base and phosphate contacts largely with the internal region of the IR previously defined as B2 while DBD2 and DBD4 interactions are located towards the external end of B2 and into A. DBD3 interacts principally with the DNA flank in a non-sequence-specific manner (Fig. Tn3.16 F bottom). There are also phosphate contacts across the IR/flank junction by residues in the catalytic RNH domain. When bound, there flank is bent from the IR axis, an observation which was expected from the enhanced [(OP)2-Cu+] cleavage sites in this region. Note the similarities with the Tn3/Tn1000 transposase organization (Fig. Tn3.16 Di).
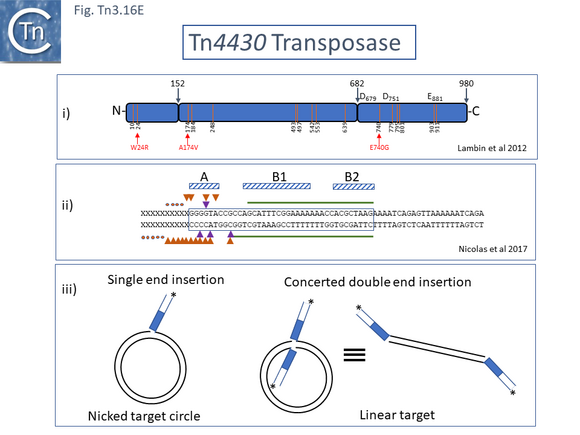
The apo-protein appears relatively compact (Fig. Tn3.16 Gi). The dimer is held together at the bottom by the DD domains and at the top by the C-terminal domain which docks onto the surface of the adjacent monomer. The CT interaction appears to be further stabilized by DNA binding (Fig. Tn3.16 Gi). The authors point out that this is an unusual dimer interface. IR binding is accompanied by large conformational change (Fig. Tn3.16 Gi). In this pre-cleavage complex, the protein “arms” align the 4 DBD along IR, bend the DNA at Site A (Fig. Tn3.16 Fiii) which moves the flank with respect to the IR tip and places the scissile phosphate bond at the catalytic site of the opposite monomer both LN and RNH residues are involved.
Like other transposition systems cleavage appears to be “in trans” (see: Cleavage in Trans: A Committed Complex), a constraint which ensures that the transpososome complex has been assembled before cleavages occur and prevents adventitious initiation of transposition. The two scissile phosphate bonds are correctly positioned to generate the expected 5bp DR. The S911 mutation which leads to hyper transposition and decreased immunity (T+/I-) would appear to assist the apo-PEC transition, as indeed would the other T+/I- mutations.
Another consequence of the transition is that, while the RNaseH fold is poorly defined in the apo-protein, it becomes more easily recognizable in the rearranged PEC. However, in this conformation only E881 (Fig. Tn3.16 Ei) is stably positioned while the other two members of the triad D679 and D751 are mobile.
The authors suggest that this is part of a regulatory process, protein metamorphism, and that additional factor(s) are involved in stabilising the catalytic pocket. It seems possibly that this may be regulated by correct docking of the target DNA. Which, they propose, could enter by opening of the DD interaction domains, a suggestion from studies with a branched DNA substrate (Fig. Tn3.16 Hi and Fig. Tn3.16 Hiii) representing a strand transfer product. The low-resolution structure suggests that the target segment of the branched molecule is located at the base (Fig. Tn3.16 Hii).
These are proposed to be the position at which the target (Fig. Tn3.16 Hiv) may dock. This led to a model of stepwise transpososome assembly in which the apo-protein first engages a target molecule which opens a “cavity” between the two protomers and subsequently allows engagement of the IR.
Tn3 Transposition immunity, a poorly understood phenomenon.
In some of the earliest studies on TnA (Tn1) [214] it was observed that transposition into a plasmid already carrying a TnA copy was severely inhibited, a phenomenon known as Transposition Immunity. The effect, identified by transposition of TnA from the E. coli chromosome to plasmid R388 or a derivative already carrying TnA was pronounced (a 105 fold reduction in the immune target).
Two other Tn3 family transposons, Tn501 and Tn1721, also exhibited this inhibition phenomenon (cited as personal communication in [215]). However, other studies have identified plasmids having received two copies of TnA but these probably occurred at the same time rather than consecutively [216][217].
Transposition Immunity is a poorly understood phenomenon and some of the early studies gave a number of conflicting results. Immunity has since been observed for bacteriophage Mu and for transposon Tn7 (e.g. [218][219][220]) where it involves proteins with ATPase activity, MuB [221][222] and TnsC [223][224] respectively. However, Tn3 and its relatives do not encode this type of protein and only a single large transposase with no demonstrated ATPase activity is involved in transposition. It is therefore possible that immunity here is mechanistically distinct from that of both phage Mu and Tn7.
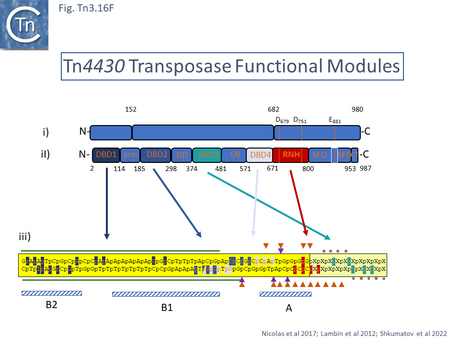
Immunity Requires a Transposon End
Further analyses of TnA [215] demonstrated that between 290 bp and 470 base pairs at the right end (Fig. Tn3.16 Ii) were sufficient to confer immunity [215]. These measurements were made either by accumulation of transposition events in bacteria grown on agar “slopes” or transpositions from the chromosome into a plasmid target in stationary phase cell [215]. While plasmids carrying the right end showed immunity, those carrying the left end showed no immunity or only “partial-immunity”.
Unfortunately, the quantitative effects are not clear from this publication. However, the conclusions are generally supported by another study which uses a different assay system involving a temperature sensitive replication mutant of plasmid pSC101 carrying a Tn3 derivative in which tnpR was inactivated by linker insertion.
In this system [176][225], cointegrates are not resolved and were isolated by “rescue” of the temperature sensitive donor plasmid by a coresident target plasmid following a shift to high temperature [226]. Here, plasmids carrying restriction fragments containing one or other Tn3 ends conferred immunity; inclusion of both ends did not enhance immunity; and immunity was observed regardless of the orientation of the 38 bp IR end. Intriguingly, the distribution of insertions into an immune and non-immune targets appeared to be different [226]. However, the study also indicated in some cases that the orientation of the Tn3 DNA fragment in the target affected the immunity level.
Furthermore, it was observed that deletions within the IRs which eliminated transposition, also eliminated immunity (Fig. Tn3.16 Iii) [227]. However, studies comparing TnpATn3 binding and immunity [187] suggested that some mutants which do not affect transposase binding capacity do impact on transposition immunity. Moreover, a study which implicated TnpRTn3 in immunity [228] was not supported by subsequent studies [227].
A finer scale analysis of the extent of the Tn3 IR sequence required for immunity was obtained by sequential deletion analysis of one IR [229] (Fig. Tn3.16 Iiii). While a number of the deletions resulted in retention of certain internal IR nucleotides, a clear pattern is that the distal end of the IR segment rather than the tip of the IR is important (sequences in Box B; Fig. Tn3.16Ci). This is also largely in agreement with the results from Huang et al.[227].
Interestingly, Bishop and Sherratt [181], using a plasmid system which allows identification of both inter- and intra-molecular Tn1 transposition Inversions and deletions were found to occur at frequencies similar to insertion suggesting that insertion into its own vector plasmid is not significantly subject to immunity. However, when Tn3 sequences, such as those present in pBR322, were also present in the transposon donor plasmid, inversions and deletions occurred at significantly lower frequencies.
For Tn1000, it was observed that 200 base pairs of the IRL (Gamma end) or 400 base pairs of the IRR (delta end) showed immunity to Tn1000 insertion [230] while no other segment of Tn1000 conferred immunity. This was further refined to the terminal 38-base-pairs of IRR which were sufficient to confer immunity, whereas the 38-bp sequence of IRL conferred only moderate immunity (note that we use the standard nomenclature for IRL and IRR: viz IRR is defined as the IR towards which the transposase is expressed. This is the opposite of the nomenclature originally used for Tn1000). The IR sequence of both ends is identical for the first 35 base pairs and it was observed that this common sequence alone was not able to confer immunity [230].
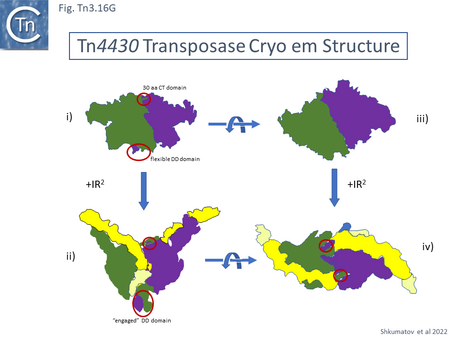
Like Tn4652 (Fig. Tn3.12 G) [110][111] in which IHF binding to sites located close to the ends positively regulates TnpA binding [111] to the terminal IRs, Tn1000 also carries IHF sites proximal to the IRs. A more detailed analysis of the related Tn1000 IRR [114][231] using a mating-out assay [232] to measure transposition frequencies, showed that while the 38 base pair end was capable of conferring immunity on a target replicon, the neighboring IHF site (which is not present in TnA/Tn1,Tn2,Tn3) conferred a significantly higher level of immunity in the presence of IHF (Fig. Tn3.16Iiv) while removal of the terminal 2 GC base pairs at the tip had no real effect. IHF has been shown to bind cooperatively with TnpATn1000 [115]. This result strongly suggested that it is the IHF-enhanced binding strength TnpATn1000 which determines the level of immunity [114].
The available data is relatively old and restricted by the experimental approaches available at that time. Since every assay system is different, it is not possible to directly compare results. However, in spite of the apparently conflicting detailed data, it appears likely that TnpATn3 and TnpATn1000 binding to an IR in the immune target is necessary for immunity.
Immunity in Tn4430
More recent studies on immunity of Tn4430 [233] have involved isolation of TnpATn4430 mutants which escape immunity [196]. The mutants were screened for both transposition and loss of immunity (T+/I-) using a papillation test. Surprisingly, these were not localized to a specific region of the protein but occurred over its entire length (Fig. Tn3.16 Ei).
The frequency of transposition into the permissive (non-immune) target of most mutants was similar to that of wild-type TnpATn4430. However, immune-deficient mutations in the N-terminal region appeared to have a slightly increased transposition frequency whereas those clustering in the C-terminus exhibited a slightly decreased transposition frequency. Based on the cryo-em structure, these T+/I- mutants are expected to positively affect the apo-PEC transition [210].
Although some data suggested that immunity could be observed in a relatively crude cell-free system [234], the establishment of a more defined and robust in vitro transposition system [193] might permit further experimental investigation into the molecular basis of Tn3 family transposition immunity.
On Ended Transposition.
Early in the study of Tn21 and Tn1721, it was observed that, In the presence of the cognate transposase, plasmids containing a single inverted repeat (IR) can fuse efficiently with other plasmids [235][236] in a reaction that requires neither the resolution system nor a functional host recA gene.
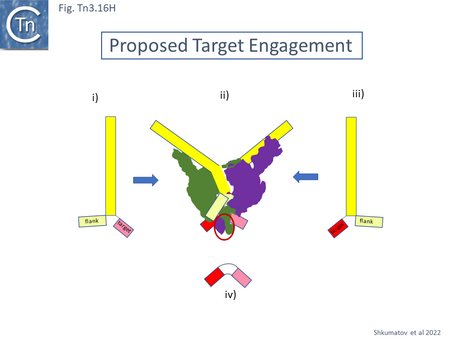
Insertion occurred at different sites in the target plasmid and the products contained a complete copy of the IR-carrying donor plasmid often with a duplication of various lengths of donor DNA. The sequence across the junction showed that the segment of donor DNA started precisely at the IR at one end, was variable at the other and the insertion was generally flanked by a 5bp DR generated in the target plasmid [237]. Some recombinants were observed to contain only short segments of the donor plasmid [238].
Models involving asymmetric (rolling circle or processive) replicative transposition or simple insertion have been proposed for this type of transposition and it seems possible that this in some way results from insertion into an extant replication fork in the target DNA.
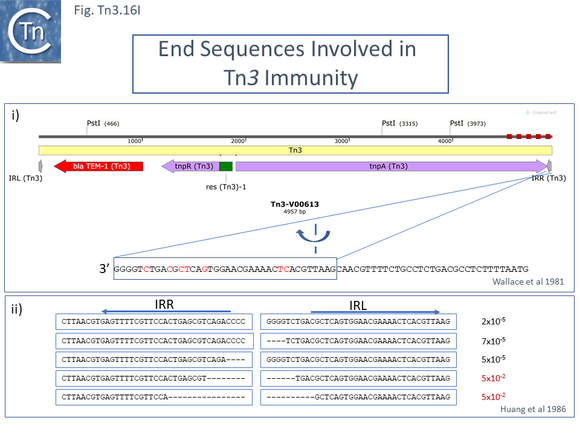
Resolution
The serine recombinases.
Efficient resolvase-catalyzed recombination between two directly repeated res sites is instrumental in completing transposition by physically separating donor and target molecules. This was first recognized in studies on complementation of transposition deficient Tn1 and Tn3 mutants where mutation of tnpR resulted in accumulation of cointegrates [8][10][225][241] (Fig. Tn3.2 ii). It therefore showed that TnpR functions not only as a repressor of TnpA and TnpR expression by binding to the res site and blocking the promoters [242] for both genes (see Fig. Tn3.17 Ci), but that it has an active function in the transposition process.
A number of resolvase enzymes have since been recognized (for a comprehensive review see [24](Fig. Tn3.17 Ai-iv).
The majority so far identified appear to be recombinases which use a serine residue as the nucleophile during recombination (Fig. Tn3.17 Ai and ii). These serine recombinases can be divided into two major groups (Fig. Tn3. 17A): the “classical” recombinases, TnpR encoded by Tn3, Tn21 and their relatives (~185 aa); and “long” serine recombinases [24][243] (~300aa) (Fig. Tn3.17 B) (see [24][244][245]. In both types, the catalytic center is located at the N-terminal end in a large catalytic domain which is followed by a smaller helix-turn-helix DNA binding domain. In the case of the “long” recombinases, there is a C-terminal extension compared to the “classic” resolvases. These fall largely within a small subclade in the Tn3 subgroup which includes Tn5044, the Xanthomonas transposons TnXc4 and TnXc5 and Tn1412 (Fig. Tn3.4A). It is worth noting that all members of this Tn group also encode a toxin/antitoxin system located between the divergent tnpA and tnpR genes (Fig. Tn3.4).
Studies with Tn1000 (γδ) and Tn3 res.
Early studies using the resolvase of Tn1000 (aka γδ) in vitro demonstrated that the enzyme could introduce double strand breaks in a res site and, in the absence of the divalent cation Mg2+, formed covalent TnpR-DNA intermediates [246].
Cleavage occurred at a crossover point in a palindromic sequence to generate a cleavage product with a free 3’OH group and a 2 base 3’ overhang [246] (Fig. Tn3.17 Ci). Furthermore, formation of a free 3’OH implied that the covalent protein-DNA linkage occurred at the 5’ end and was more efficient if the substrate carried 2 directly repeated res copies. This led to the hypothesis that although TnpR acts as a repressor at res, binding simultaneously to two res copies in some way changes the protein conformation allowing recombination to proceed [246]. It was further shown using DNase and footprinting that resTn3 and resTn1000 carry three TnpR binding sites [247], I, II and III (where sites II and III, known as accessory sites, are closely spaced and site I known as the core site, is very slightly distanced) (Fig. Tn3.17 Ci) [247] and that the recombination point (the dinucleotide TA) [248] is included within site I. Each site shows some degree of two-fold symmetry [247][249] (Fig. Tn3.17 Ci). The resTn3 has an identical organization [250] and almost identical sequence and the Tn3 and Tn1000 TnpR products are interchangeable [250]. These similarities were exploited to determine the crossover point using Tn1000 TnpR-mediated resolution between resTn3 and resTn1000 carried by a single plasmid [248].
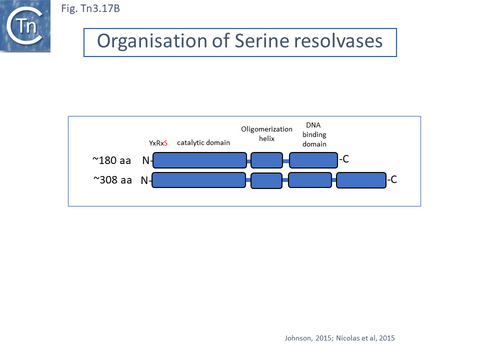
Tn3 res, tnpR and tnpA gene expression.
In both Tn3 and Tn1000, tnpA and tnpR are divergent and the res site is located in the intergenic space with subsite III proximal to tnpR (Fig. Tn3.17 Ci). Promoters for both tnpA and tnpR, were located by footprinting of RNA polymerase and lie within res [242][252] (Fig. Tn3.17 Ci). The -35 promoter elements of both gene are only 10 bp distant from each other and the -10 element of tnpA is located within site I straddling the point of recombination crossover (Fig. Tn3.17 Ci). The transcription start point for both genes has been mapped. Clearly, tnpA and tnpR expression would be regulated by TnpR binding.
Variant res sites with this configuration have been observed in which the center of sites I and II are separated by 4, 5, 6 and seven helical turns (see [24]).
The Mechanics of Resolution.
TnpR binding to res generates a highly compact protein-DNA complex as judged by electron microscopy [253]. This was explained by the observation that TnpR binding to res-containing linear DNA fragments results in significant bending of the DNA although it was noted that the complex contains a single DNA molecule under the conditions use rather than two res sites [254].
Gentle proteolysis of purified Tn1000 TnpR was observed to generate two fragments: a large N-terminal fragment which includes the catalytic center and a smaller C-terminal fragment which binds to each of the three res sites [255](Fig. Tn3.17 B). Unlike full length TnpR which binds the res sub-sites with equal affinity, the C-terminal fragment binds to each of the half-sites but with different affinities suggesting that the N-terminal part of TnpR is involved in protein-protein interactions within the TnpR-res complex [255]. Footprinting of small fragment binding indicated that the protection was centered on the 9bp half-sites (Fig. Tn3.17 Ci). Further studies using saturated mutagenesis of a halfsite from subsite I and chemical probing identified how the protein contacts DNA in both the major and minor grooves [256].
A model of the overall architecture of single TnpR-res complexes was proposed [254] based on results using a number of footprinting agents to reveal sensitive sites on the DNA and permutation experiments to identify DNA curvature [257] in which each subsite binds a TnpR dimer (with one monomer recognizing each partial diad symmetry element called “half-sites”) [247][249] and introduces an “intra-site” bend in the DNA at each site while at another level, protein-protein interactions introduce inter-site bends (Fig. Tn3.17 D). Experimentally, this conformation requires all 3 sites and a correct spacing between sites I and II.
In vitro resolution systems have been developed and require supercoiled DNA together with a divalent cation, Mg2+ [30][246][250][258]. A number of laboratories have contributed to an understanding of how the complex site-specific resolution recombination reaction takes place. These studies have used extremely clever techniques to understand the mechanics of this process including topology, mutagenesis and structural biology.
In vitro resolution requires a supercoiled DNA substrate carrying two directly repeated res sites and results in a simple concatenated recombination product with a specific change in linking number (the number of time one DNA strand crosses another) (Fig. Tn3.17 E) [30][246][250][258] indicating that the synaptic complex must have a very precise type of protein-DNA architecture. The in vitro reaction is very inefficient when the res sites are in an inverted orientation raising the question of how the two res sites are aligned for recombination (for review see [29]). Random collision between res sites on a supercoiled molecule was ruled out since this would generate a complex concatenated product with a variable number of supercoils trapped between the recombined product (Fig. Tn3.17 Eii). Alignment of the two res sites was first proposed to occur when TnpR recognizes one site and tracks along the DNA molecule until encountering the second site. However, present evidence suggests that this is not the case [123]. In particular the observation that res site recombination can occur intermolecularly. A second hypothesis was that res sites meet via “slithering” i.e. continuous one-dimensional diffusion of supercoils in plectonemically (Fig. Tn3.17 Eii) wound DNA molecules (for review see [29]).
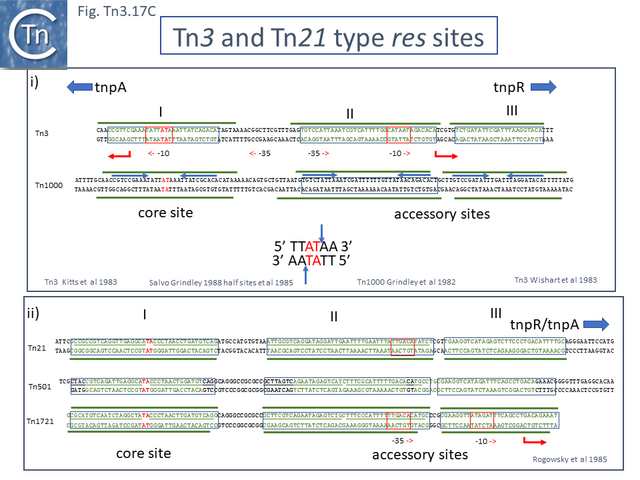
Intensive studies using both gel electrophoresis and electron microscopy to visualize TnpR recombination activities [261][262] led to a model in which the two res Fig. Tn3.17 Eiii) sites are constrained in a configuration which entraps 3 supercoils (Fig. Tn3.17 Eiiib) and which takes into account the observation that Tn3 resolution (Fig. Tn3.17 Eiiic) removes four negative supercoils on recombination (Fig. Tn3.17 Eiiid) [30]. The resulting energy change probably drives the reaction. In this model, it is TnpR interactions at the accessory sites II and III which are important for this allowing the recombining site I to finalize the recombination event (Fig. Tn3.17F).
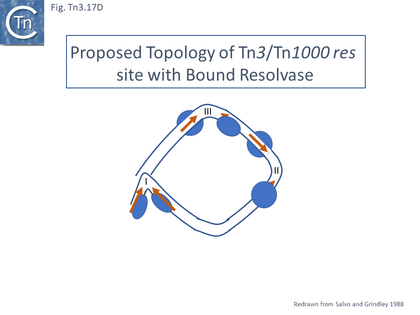
This occurs by simple rotation at site I [29] on the flat hydrophobic surface after simultaneous cleavage of all four strands in the synaptic complex. The TnpR monomers remain attached to the 5’ ends (Fig. Tn3.17 F left) and the serine-DNA bond is then broken by attack by the 3’OH of the recombining site (Fig. Tn3.17 F right) to complete recombination. More than a single round of recombination can occur and this results in the generation of knots of increasing complexity with increasing numbers of recombination events (not shown).
This model is supported by the structure of a TnpR tetramer bound to two site I DNA molecules in a synaptic complex [263][264] which shows that each TnpR dimer bound to its DNA presents an unusual flat, hydrophobic surface to the other member of the pair (Fig. Tn3.17 G) with the suggestion that strand exchange indeed occurs by rotation around this interface.
The Tn3 synaptosome
An understanding the structure of the Tn1000/Tn3 synaptosome is important to validate the proposed resolution reaction pathway (Fig. Tn3.17 E). Visualization of the entire synaptosome has proved challenging due to its complexity: with two res sites each composed of three TnpR binding sites, the catalytic site I and the “architectural” sites II and III, each with its six TnpR dimers. The first synaptosome structure obtained was that of Sin, a simpler organization with only two TnpR binding sites [243][265][266]. A long awaited Tn3 synaptosome complex has now been described at the structural level [267] (Fig. Tn3.17 H) and provides convincing evidence for the previous models (Fig. Tn3.17 Eiii).
The Tn1721, Tn21 and Tn501 res.
In contrast to those of Tn3 and Tn1000, the tnpA and tnpR genes of Tn1721, Tn21 and Tn501 are transcribed in the same orientation, with tnpR upstream of tnpA and their res sites located upstream of tnpR (Fig. Tn3.17 Aii and Fig. Tn3.17 Cii). They are relatively well conserved within the Tn21 clade (Fig. Tn3.7 F). Early experiments with Tn501 showed that it too underwent transposition using a cointegrate intermediate [268].
Like resTn3 and resTn1000, resTn1721 and resTn21 are composed of three TnpR binding sites (I, II and III) as determined by footprinting [269][270] with site III proximal to tnpR (Fig. Tn3.17 Aii and Fig. Tn3.17 Cii) and each site exhibits some degree of dyad symmetry. Moreover, there is considerable identity observed the Tn21, Tn501 and Tn1721 tnpR genes and also between the resTn21 , resTn501 and resTn1721 sites [35].
All three elements complement a tnpR mutant of Tn21 whereas Tn3 does not [35]. This is perhaps not surprising since the resTn3 sequence appeared to be quite different from those of this Tn group (Fig. Tn3.17 C) and the authors were unable to identify a res site homologous to that of Tn3. In addition, the TnpR amino acid sequence of Tn3 is somewhat distant from those of Tn21, Tn501 and Tn1721.
These observations were reinforced by additional studies demonstrating that purified Tn1721 TnpR can resolve cointegrate substrates containing repeat copies of resTn1721, of resTn21, and of a substrate carrying both resTn21 and resTn1721 copies, but not of resTn3 [271] while Tn21 TnpR catalyzed site-specific recombination between directly repeated resTn21 and resTn1721 but not resTn3 [272]. The reaction required a supercoiled substrate with two directly oriented res sites.
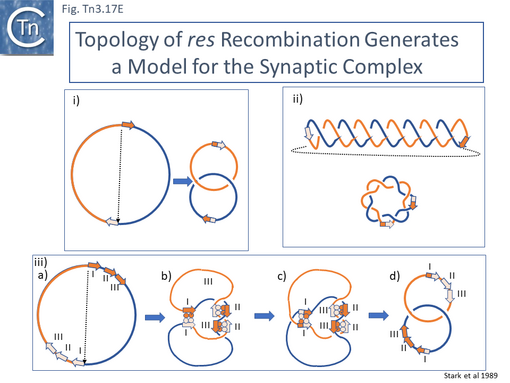
Several studies explored the DNA sequence binding and recombination specificities between Tn3 and Tn21 using hybrid TnpR containing the DNA binding domain of one and the catalytic domain of the other [273][274][275]. These studies showed that, while a Tn21 TnpR catalytic DNA domain spliced to the Tn3 DNA binding domain has a somewhat lower affinity for resTn21, it retained some ability to mediate recombination between resTn21 but was unable to recombine resTn3 sites in spite of the fact that the hybrid protein was able to bind resTn3. This led to the conclusion that although
“alterations in amino acid sequence of resolvase within the helix-turn-helix DNA binding domain modulate the affinity of the protein for its DNA target sequence, the specificity of resolvase for recombination at its cognate res sites is determined by the resultant organization of the DNA-protein complex” [274].
Tn res activity tnpR and tnpA gene expression.
It was proposed [35] that in all three elements, tnpA may be transcribed independently of tnpR and that its promoter is located within tnpR. Moreover, no Tn501 tnpR promoter could be found in vitro. This is consistent with the observation that in interreplicon Tn501 transposition into plasmid R388, resolution could be induced in the recipient by mercury selection [268] suggesting that tnpR may be expressed at least partially as part of the mercury resistance operon located upstream of tnpR.
Interestingly, a study using Tn21 revealed a gene, tnpM (for modulator), whose expression appeared to enhance transposition and suppress resolution [64]. TnpM results from the insertion of the Tn402 derivative, Tn5060 which led to the formation of Tn21 (Fig. Tn3.7 G). This event interrupted the urfM gene, of unknown function but possibly part of the mercury operon, generating the C-terminal fragment with a fortuitous translation initiation codon. Removal of the region in Tn21 resulted in a reduced transposition frequency and increased resolution activity and these activities were restored when the tnpM “gene” cloned into another compatible plasmid was provided in trans. Moreover, transposition of Tn501, which like Tn21, also includes a complete ufrM gene, was also affected. The mechanism by which the UfrM fragment, TnpM, functions is unclear and has not been addressed since its initial description [64].
The long serine recombinases
TnpR proteins carrying an extended C-terminus (TnpRL) (Fig. Tn3.17 B) have been studied in only a single case, TnXca5 (ISXca5) from Xanthomonas campestris pv. citri XAS450 [276]. Establishment of an in vitro system [277] has shown that, as for the short forms of TnpR, recombination requires two directly repeated resTnXc5 copies in a supercoiled plasmid substrate and Mg2+ as a divalent cation. Footprinting reveal three TnpR-binding subsites with a relative spacing and similar position with respect to tnpRL as Tn3 and Tn1000 (Fig. Tn3.17 I).
Topological analysis of the recombination products suggests that the resTnXc5 synaptic complex must be very similar to those of resTn3 and resTn1000 since 4 supercoils are lost on recombination and the directionality of strand exchange is the same [277]. No structural studies are at present available.
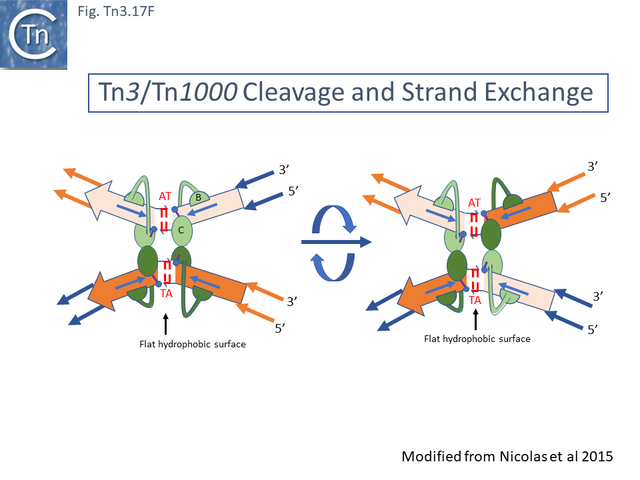
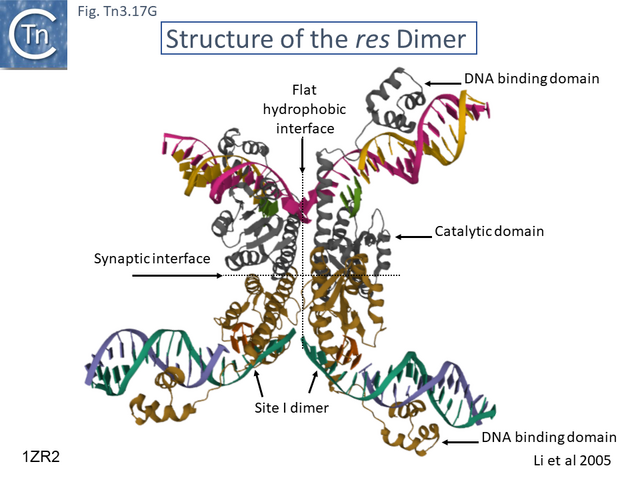
Serine-recombinases which use IHF/Hu: the Sin Synaptosome.
It is worth noting that certain serine recombinases, such as Gin and Hin , involved in inversion switches [245][278][279][280] or Sin which is involved in plasmid recombination [243][281][282] use “simpler” recombination sites but depend on DNA bending proteins such as IHF, Fis, HU and HUB to achieve the correct architecture. These are not known to act in the resolution process of Tn3 family transposons.
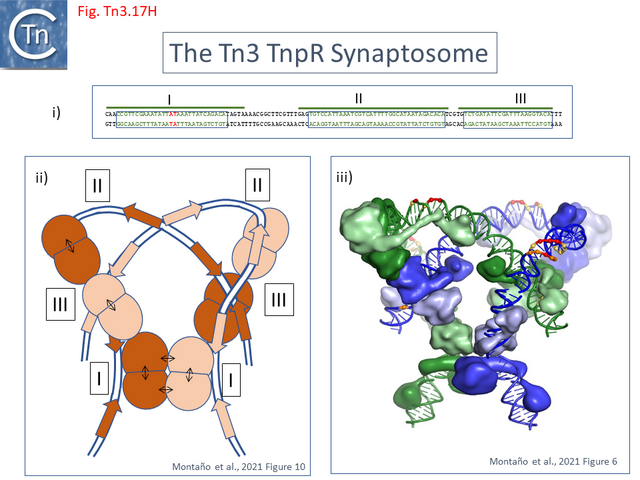
The Staphylococcal Sin resolvase was identified as part of a multi-resistance staphylococcal plasmid, pI9789 [284]. While Sin is not part of a transposon or an invertible DNA segment, DNA sequences immediately upstream of the Sin binding site, sin, are favored integration sites for Tn554-family transposons. The sin site is composed of only two Sin binding sites I and II and an intervening region bound by an HU-like DNA bending protein such as the Bacillus subtilis Hbsu (Fig. Tn3.17 Ii). The functional site includes four imperfect: two inverted copies of a 12-bp motif repeat (sites I-L and I-R) and two in direct repeat (sites IIa and IIb) [243]. While Sin on its own binds to sites I and II to generate DNase-protected regions, Hsbu on its own showed no protection. However, together, the two proteins result in protection of the entire site (note that protection is not even and additional sites of hypersensitivity are generated [243].

Similar biochemical and topological and genetic approaches to those used in the analysis of Tn3/Tn1000 TnpR/res function were used to investigate the Sin/sin system [265][281]. This showed that the synaptosome traps three supercoils between the points of strand exchange and requires both site I, where strand exchange occurs, and site II together with Hbsu. A synaptosome structure, the first determined foe a resolution system, has been solved [265][285] and a cartoon showing the arrangement of Sin dimers and a dimer of dimers bridging the two sin sites is shown in Fig. Tn3.17 J. This illustrates how bound non sequence-specific DNA bending proteins assist in synaptosome formation.
The irs/TnpI system
A small group of known Tn3 family members which include the Bacillus thuringiensis transposons Tn4430 [286] and Tn5401 [287] encode a resolvase, TnpI, carrying a tyrosine residue at the active-site nucleophile [31][287][288] (Fig. Tn3.17 Ki). The tnpI gene lies upstream of tnpA and both genes are transcribed in the same direction (Fig. Tn3.17 Aiii). Insertion mutagenesis showed that interruption of tnpI resulted in an increased level of cointegrate intermediates in Escherichia coli [31].
The Tn4430 sequence [31] revealed a series of small sequence repeats directly upstream of tnpI as well as two smaller repeats abutting the inside border of IRL. The tnpI proximal repeat sequences include two 14bp inverted repeats, IR1 and IR2, together with two longer direct repeats, DR1 and DR2, related in sequence to IR1 and IR2 (Fig. Tn3.17 Kii).
DNase footprinting revealed that TnpI bound to all four sites together called the internal resolution site (irs) [174] but not to the (unrelated) IRL proximal repeats (Fig. Tn3.17 Kii). Using a suicide substrate which contains a nick close to the point of recombination and which traps intermediates in the cleavage reaction, in an in vitro reaction TnpI was found to be able to bind to a linear DNA fragment containing IR1-IR2 and did not require assistance from the two DR repeats.
DNA cleavage is staggered occurring six base pairs apart [174] (Fig. Tn3.17 Kii) forming a transient 3′-phosphotyrosyl bond leading to 3’OH in an identical way to other tyrosine recombinases (e.g. [289][290][291][292][293][294]. A complete in vitro resolution reaction requires supercoiled DNA substrate [174].
A similar overall sequence architecture was observed upstream of tnpI in Tn5401 [288] (Fig. Tn3.17 Kii). Here, the repeated sequences are 12bp long with identical repeats abutting the inside border of IRL and of IRR. Footprinting also identified the TnpI irs binding sites but, in addition showed TnpI binding to the IR proximal site [288].
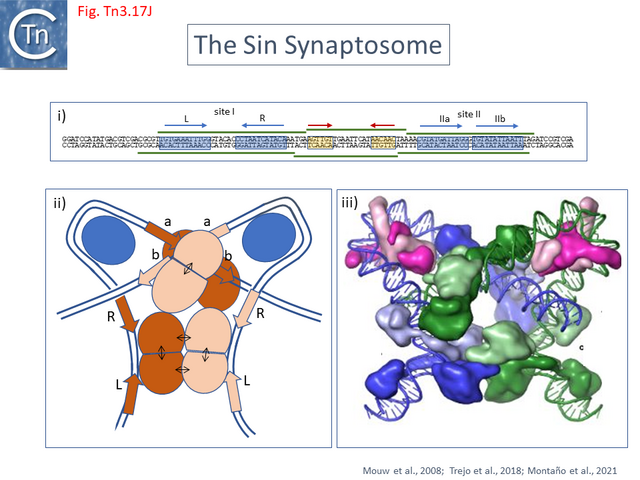
The Mechanics of Resolution.
In contrast to the requirements for the accessory sites I and II in serine recombinase-catalyzed resolution [257], there is no absolute requirement for the DR1 and DR2 accessory sites for activity in TnpI-catalyzed recombination. Instead, in their absence IR1-IR2 core site recombination can give rise to different recombination products such as deletions, inversions and intermolecular recombination in vivo and topologically complex products in vitro instead of the simple catenanes [174]. In other words, the accessory sites channel recombination to generate resolutive recombination between two directly repeated irs sites on the same DNA molecule. This gave rise to the model shown in Fig. Tn3.17 K.
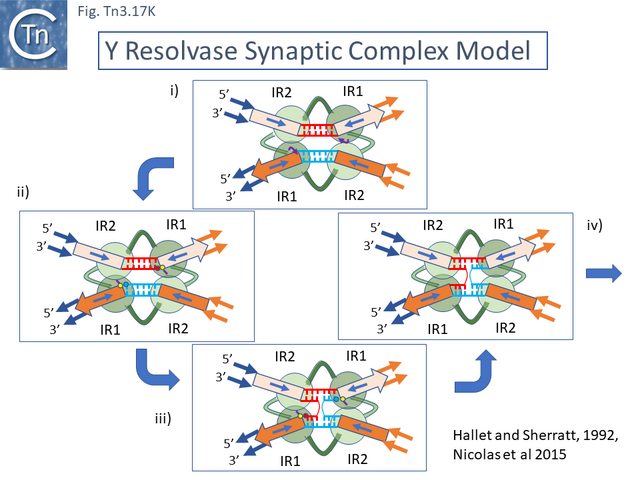
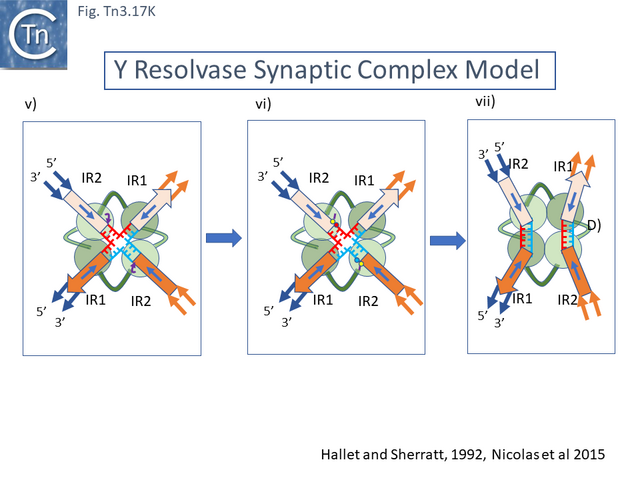
More specifically, formation of synapses including DR1 and DR2 was found to stabilize recombination intermediates favoring the forward recombination reaction and to impose an order of cleavage at the IR1-IR2 core sites: activation of the IR1-bound TnpI subunits (those furthest from the accessory sites) occurs resulting in IR1 cleavage (Fig. Tn3.17 Lii) and first strand exchange Fig. Tn3.17 Liii to form a Holliday junction (Fig. Tn3.17 Liv) while the second pair, the IR2-bound subunits, are then activated to resolve the holliday junction Fig. Tn3.17 Lv) by cleavage and exchange of the second strand (Fig. Tn3.17 Lvi) to resolve the cointegrate Fig. Tn3.17 Lvii) [174][297].
Although the exact topology of the synaptic complex is unknown, two alternative models [24] lead to the conclusion formation of the synaptic complex induces the same net change in substrate topology, trapping two negative supercoils between the crossover sites and converting them into catenation nodes in the product (see Fig. Tn3.17 L).
Irs, tnpR and tnpA and gene expression.
Transcriptional start sites within Tn5401 were mapped by primer extension analysis and the -35 and -10 promoter elements were identified (Fig. Tn3.17 Kiii) [287]. Two overlapping and divergent promoters were identified: one which would drive expression of tnpI and tnpA and the other which could drive the upstream but divergent toxin antitoxin genes (see: Tn3 family-associated TA passenger genes are located in a unique position).
The rst/TnpS/T system.
The third major type of resolution system encoded by Tn3 family members is the TnpT-TnpS system which uses a resolution site, rst (res site for TnpS and TnpT) encoded by the catabolic transposons Tn4651 [298]. Tn4651, isolated from a Pseudomonas plasmid carries a set of toluene degrading (xyl) passenger genes (Fig. Tn3.3) and is similar to the mercury resistance transposon Tn5041 (Fig. Tn3.17Ki) [117]. The tnpS and T genes are expressed divergently with the res site between the two. In some cases, tnpT and tnpS are separated by insertion of passenger genes (Fig. Tn3.17 Lii). Resolution of cointegrates generated by Tn4651 was shown to require three Tn4651-encoded factors: the res site (now called rst) and the tnpS and tnpT gene products which are located at a significant distance (48kb) away from the tnpA transposase gene. A similar long distance between transposase and resolvase is found in Tn5041 [118] Fig. Tn3.17 K). Here, tnpS and tnpT are referred to as orfQ and orfI respectively and rst as attTn5041.
The 323 aa TnpS protein is a tyrosine recombinase (Fig. Tn3.17 O) with similarity to the Cre resolvase [24] while the 332 aa TnpT appears to enhance TnpS-mediated recombination [32].
The sequence of the tnpS/T intergenic region is very similar in Tn4651[174] , Tn4652 (a Tn4651 deletion derivative lacking the toluene-catabolic genes) [300], Tn4661 [28] and Tn4676 [119]. It is composed of a 203 bp sequence which includes two pairs on inverted repeats, IRL and IRR and IR1 and IR2 (Fig. Tn3.17 Pi) with overlapping promoters which drive TnpS and TnpT expression. The mRNA start point was identified by primer extension [32].
The length of functional rst site, 136 bp, was defined by the recombination activities of sequential deletion derivatives in an in vivo resolution system [24]. This involved the construction of an artificial cointegrate containing one complete rst copy and a second copy which carries the deletions. IR1 and IR2 are indispensable for the full resolution activity and cointegrate resolution was shown to require both TnpS and TnpT.
Moreover, the resolution reaction could be reversed to obtain site-specific integration (recombination between rst sites on different DNA molecules) in a reaction which requires TnpS but not TnpT. Suggesting that TnpT is a factor which determines the direction of recombination [24][32].
Although the TnpS/T proteins of Tn4651 and Tn4661 are highly similar and the Tn share highly similar sequences in the inverted repeat motif, IRL and IRR, of the rst core site, the 7bp spacer separating the repeats are somewhat different (Fig. Tn3.17 Pi). An artificial cointegrate composed of an rstTn4651 and an rstTn4661 site could not be resolved using TnpSTn4651 and TnpTTn4651 [28]. The mismatches in the IRL-IRL region concern principally the spacer region between IRL and IRR. rstTn4651 and rstTn4661 have six mismatches, with five located in the spacer (Fig. Tn3.17 Pii). In other tyrosine recombinase systems such as xerC/D or phage P1 Cre protein (see [24][289][301]) this is the region where strand cleavages occur and sequence differences have a strong influence on the ability for two sites to recombine.
The effect of these sequence differences was investigated using a cointegrate, in which the spacer sequence of rstTn4661 was replaced with that from rstTn4651, (rstTn4661v1) (Fig. Tn3.17 Nii). In contrast to the cointegrate carrying both rstTn4651 and rstTn4661, that carrying rstTn4651 and rstTn4661v1 underwent TnpSTn4651 and TnpTTn4651-mediated resolution, demonstrating that it is the differences spacer sequence which prevents recombination [28]. Moreover, the sequence of this region in the resolved products confirmed that recombination occurred within the IRL-IRR region. It should be noted that neither the tnpS/T intergenic sequence nor the proposed core recombination site of Tn5041 [117][118] (Fig. Tn3.17 K) shows significant similarity to those of Tn4661 [28]. Moreover, in depth analysis of the Tn5041 resolution reaction has not been reported.
No footprinting, binding stoichiometry or topology studies are yet available for the TnpS/T system and the exact role of TnpT in the resolution reaction is not known although it has been demonstrated to bind the DNA region containing IR1 and IR2 [28].
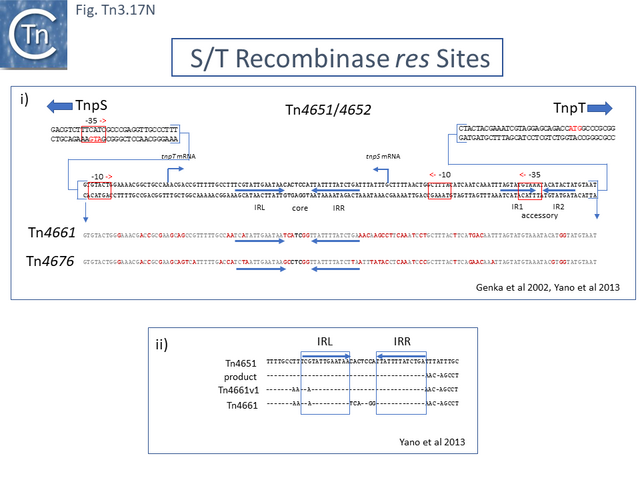
Toxin-Antitoxin genes: Special Passengers linked to the transposition process?
Several studies had identified individual Tn3 family members with type II toxin-antitoxin (TA) passenger genes [34][143][303][304] (see reference [24]). Unusually for passenger genes of this Tn family, the T/A genes are consistently found adjacent to a resolvase gene.
Some type II TA systems are involved in plasmid maintenance in growing bacterial populations by a mechanism known as post segregational killing. Upon plasmid loss, degradation of the labile antitoxin liberates the toxin from the inactive complex, which in turn is free to interact with its target and cause cell death. They were first identified in the mid-1980s in plasmids F [305] and R1 [306] and it was recently shown that acquisition of a Tn3 family transposon Tn6231 carrying a type II TA gene pair was indeed able to “stabilize” an unstable target plasmid [304].
Many different type II TA gene pairs have now been identified in bacterial chromosomes as well as plasmids [307][308][309]. They are generally composed of 2 relatively short proteins: a stable toxin and a labile antitoxin that binds the toxin and inhibits its lethal activity (see reference [308]). The antitoxin includes a DNA binding domain involved in promoter binding and negative regulation of TA expression.
Identification of TA gene pairs in Tn3 family members.
Among nearly 200 Tn3 family members, 39 were observed to carry type II T/A genes (colored squares; Fig. Tn3.4A, Fig. Tn3.18 B) [34]. The host transposons included examples from all known combinations and orientations of transposase and resolvase genes (Fig. Tn3.18 A) and were almost all located adjacent to the resolvase genes in family members with TnpR, with long serine TnpR, with TnpI (e.g. Tn5401 and TnBth4) and with TnpS/T (e.g. TnHdN1.1) (Fig. Tn3.4A, Fig. Tn3.18 B). Illustrative examples are shown in (Fig. Tn3.18 A).
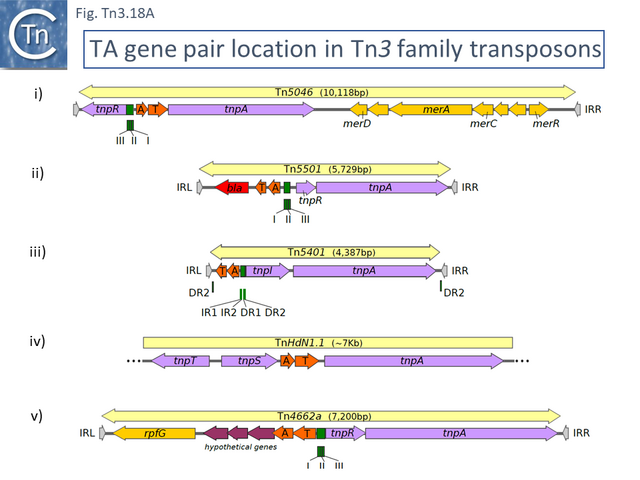
TA diversity in Tn3 family members.
The Tn3-associated TA modules include a number of different types of TA module: 5 toxin (RelE/ParE, Gp49, PIN_3, PIN,and HEPN) and 6 antitoxin families (ParD, HTH_37, RHH_6, Phd/YefM, AbrB/MazE, and MNT) (Fig. Tn3.4A; Fig. Tn3.18 B). All, except ParE, are associated with RNase activity [307][308][310], while ParE inhibits DNA gyrase activity by an unknown molecular mechanism [311].
TA distribution and organization within the Tn3 family
The majority of examples occurred in two Tn3 subgroups: Tn3 (2 toxin families) and Tn3000 (3 toxin families), but other subgroups also included members with T/A modules (6 members of 5 different toxin families (ParE, Gp49, PIN_3, PIN, and HEPN). The majority of T/A-containing members of the Tn3 subgroup also encode a long serine recombinase, TnpRL as their resolvase and two (Tn5401 and TnBth4) encode the tyrosine TnpI resolvase, while those in the Tn3000 subgroup all encode a short serine resolvase, TnpRS (Fig. Tn3.4A; Fig. Tn3.18 B). It is also noteworthy that, a given toxin gene can be paired with different antitoxins forming 7 different toxin-antitoxin pairs: ParE-ParD, ParE-PhD, PIN_3-RHH_6 (??), Gp49-HTH_37, PIN-Phd, PIN-AbrB, and HEPN-MNT (Fig. Tn3.18 B).
Although TA genes are generally arranged with the antitoxin upstream of the toxin gene, TA systems of reverse order have been identified [308]. Among the Tn3 family-associated TA systems, in five Tn3000 subgroup members (Fig. Tn3.18 B and Fig. Tn3.4A) the RelE/ParE superfamily toxin Gp49 (PF05973) toxin gene precedes that of a HigA superfamily antitoxin, HTH_37 (PF13744) [307][308][309]. A similar situation is found in the unrelated Tn4651 subgroup member TnPosp1_p.
In addition to encoding a TnpI resolvase, Tn5401 and TnBth4, both encode a ParD antitoxin, which appears to lack the DNA-binding domain.
Acquisition and exchange of TA modules.
An important question is whether these systems have been repeatedly recruited or have evolved from a common ancestor. Putting aside the fact that several groups of T/A encoding Tn (e.g. Tn5051 and its derivatives which differ essentially by their other passenger genes), clearly the fact the Tn collection also includes examples of different combinations of T/A genes and examples in which the gene order has been inverted argue for a certain level of repeated acquisition.
In cases where the TA module is found in related transposons (with similar tnpA and/or resolvase genes), it is likely that it was first acquired by a transposon that subsequently diverged. Alternatively, for transposons which are generally not related (different tnpA family group, different resolvase) but which harbor TA modules that are similar at the DNA level, it is likely that the TA module was acquired by recombination with another transposon.
Phylogenetic analysis suggested that ParE had been acquired three times, Gp49 together with an HTH antitoxin on three occasions and PIN on two occasions [34].
Although it is unclear how most of the T/A modules were initially acquired, it is important to underline that the res/rst/irs sites are highly recombinogenic in the presence of their cognitive resolvases producing transitory single (Y-recombinases) or double (s-recombinases) breaks. It seems possible that the modules were recruited via non-productive resolution events.
Moreover, this recombination activity has clearly led to spread of T/A modules to different Tn3 family members by inter-res recombination (Fig. Tn3.18 B). There are two cases in the Tn3 library, both involving Tn5051 and its derivatives, which demonstrate this capacity (Fig. Tn3.18 C). In the first case (Fig. Tn3.18 Ci), comparison between Tn5051 and TnTsp1 shows a clear break in the homology between the two Tn which occurs at the res site (Fig. Tn3.18 Ci). The identities towards the right of the res site III decrease rapidly within a short distance. In the second case (Fig. Tn3.18 Cii), comparison of Tn5051 and Tn4662a shows a clear break in identity at res site I suggesting that they have previously exchanged left and right ends via recombination at res site I. An additional transposon, Tn5051.12 is clearly a hybrid of these two since it carries the left end of Tn4662a and the right end of Tn5051.
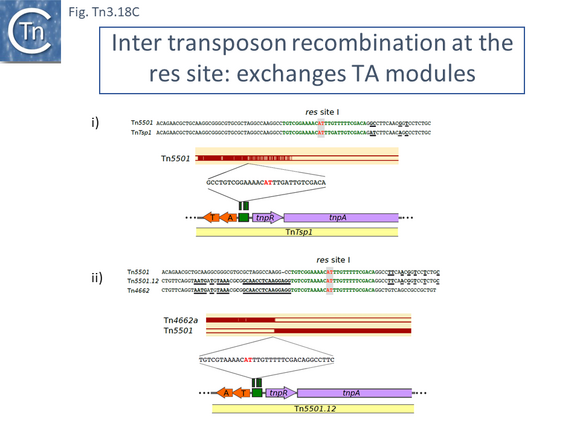
Tn3 family-associated TA passenger gene are located in a unique position.
In most of the cases identified, the T/A modules are embedded within the transposition module comprising transposase and resolvase genes and the res site at a position very close to the res sites (Fig. Tn3.18 A). This is in sharp contrast to all other Tn3 family passenger genes, which are generally located away from the resolution and transposon genes and, where known, have often been acquired as integron cassettes or by insertion of other transposons.
Indeed, several TA-carrying transposons represent closely related derivatives with identical transposase, resolvase, and TA modules but contain different sets of passenger genes (e.g., Tn5501.1 and derivatives 5501.2, 5501.3, 5501.4, etc.). Most T/A modules [34] are located directly upstream of the resolvase genes (tnpR, tnpRL or tnpI) (Fig. Tn3.18 A) with only three exceptions: a single example of a derivative with the TnpS/TnpT resolvase, TnHdN1.1 (Fig. Tn3.18 Aiv), where they are located between the resolvase tnpS and transposase genes; TnSku1 [Tn7197], where they are located downstream of and transcribed towards tnpR; and a partial transposon copy, TnAmu2_p with a short open reading frame (ORF) of unknown function between the divergently transcribed antitoxin and tnpR genes.
Regulation of Tn3 family TA gene expression.
An as yet unanswered question is how expression of the identified Tn3-associated T/A genes is regulated. It is possible that it occurs from their own promoters although it has not yet been demonstrated any of the T/A modules carry their own promoters. Alternatively, the fact that the genes are embedded in the transposition modules, it is tempting to speculate that they may be regulated in a similar way to tnpR and tnpA expression.
Tn3 family with TnpR and TnpRL
In Tn3 itself, which has been examined in detail, transposase and resolvase gene expression is controlled by promoters found within the res site located between the two divergent genes (Fig. Tn3.17C) which are regulated by resolvase binding. The location of the TA genes in proximity to the res sites raises the possibility that their expression is also controlled by these promoters (Fig. Tn3.18 Di). Although few of the res sites in the collection of TA-associated Tn3 family members have been defined either experimentally or by sequence comparison, 27 potential sites were identified [34] using the canonical tnpR-associated res-site organization schematized in [24] as a guide, a res site library (kindly provided by Martin Boocock), and RSAT tools (Regulatory Sequence Analysis Tools) [34]. For transposons with a TnpR or TnpRL resolvase, the TA genes are always located just downstream from res site I, whereas tnpR is located next to res site III (Fig. Tn3.18 A i, ii and v; Fig. Tn3.18 Di).
In transposons with divergent tnpA and tnpR such as TnXc5 and Tn5563a (Fig. Tn3.18 Di), tnpA, tnpR and res are organized similarly to those of Tn3, which itself does not carry the TA module, except that tnpA is separated from res by the intervening TA genes. This organization is also similar in Tn3 members in which tnpA is downstream of tnpR and in the same orientation (e.g., Tn5501 and Tn4662a; Fig. Tn3.18 A i and v).
Tn3 family with TnpI
Promoters have also been defined in the res (irs) site of the tnpI-carrying Tn5401 [287][288][312], and tnpI and tnpA expression is modulated by TnpI binding to the irs site [312] (Fig. Tn3.17I). The other tnpI-carrying transposon with TA genes, TnBth4, has an identical irs site, and therefore expression is probably regulated in the same way. Again, the potential promoters are pertinently located for driving expression of the TA module (Fig. Tn3.18 Dii).
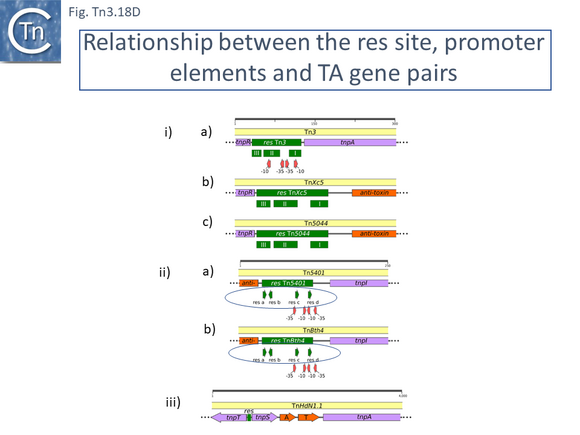
Tn3 family with TnpS/T
Finally, transposon TnHdN1.1 (Fig. Tn3.18 Aiv) is the only example in our collection of a tnpS/tnpT transposon carrying a TA module. The res (rst) site and relevant promoter elements for the divergently expressed tnpS and tnpT have been identified between the two genes in transposon Tn4651 (Fig. Tn3.18 Diii). In TnHdN1.1, the TA gene pair is to the right of tnpS, between tnpS and tnpA, and all three genes are oriented in the same direction. Although the exact regulatory arrangement remains to be determined, it seems possible that the promoters in the rst site regulate expression of the TA gene pair.
Thus, for all three types of resolvase-carrying Tn3 family members, the T/A gene module is strategically placed so that it could be place under control of the resolvase/transposase transcriptional expression signals except for the two exceptions TnSku1 and TnAmu2_p. T/A activity could therefore be intimately linked to the transposition process itself rather than, or in addition to, simply providing a general addiction system that stabilizes the host replicon, generally a plasmid, carrying the transposon.
A model for T/A activity in transposon transposition.
Type II TA expression, like that of tnpA and tnpR, is tightly regulated at the transcriptional level (see [308]). Where analyzed, the toxin-antitoxin complex binds via the antitoxin DNA-binding domain to palindromic sequences located in the operon promoter and acts as a negative transcriptional regulator. This regulation depends critically on the relative levels of toxin and antitoxin in a process known as conditional cooperativity, a common mechanism of transcriptional regulation of prokaryotic type II toxin-antitoxin operons in which, at low toxin/antitoxin ratios, the toxin acts as a corepressor together with the antitoxin. At higher ratios, the toxin behaves as a derepressor. It will be important to determine whether the Tn-associated TA genes include their indigenous promoters [308][313][314].
Transposon Tn6231 [304] (99% identical to Tn4662) clearly provides a level of stabilization of its host plasmid implying that TA expression occurs in the absence of transposition. There are a number of ways in which this could take place (Fig. Tn3.18 E). Expression could occur from a resident TA promoter (Fig. Tn3.18 Ei) if present. However, this might lead to expression of the downstream tnpA gene by readthrough transcription. Alternatively, in the absence of a TA promoter, TA expression could occur stochastically from the res promoter (Fig. Tn3.18 Eii). However, this does not rule out the possibility that TA expression is regulated at two levels with a low-level “maintenance” expression, resulting in the plasmid stabilization properties described by Loftie-Eaton et al. [304] together with additional expression linked to derepression of the tnpA (and tnpR) promoters that must occur during transposition (Fig. Tn3.18 Eiii). Regulation of tnpR and tnpA by TnpR is a mechanism allowing a burst of TnpA (and TnpR) synthesis, transitorily promoting transposition as the transposon invades a new host. Subsequent repression by newly synthesized TnpR would reduce transposition activity, reinstalling homeostasis once the transposon has been established, a process similar to zygotic induction [315] or plasmid transfer derepression as originally observed for the plasmid ColI [316] and subsequently for plasmids R100 [317] and R1 [318]. An alternative but nonexclusive explanation stems from the observed enhanced plasmid stability afforded by Tn6231 TnpR, in addition to that afforded by the neighboring TA system [304]. Resolvase systems are known to promote resolution of plasmid dimers (see reference [43]), and it was suggested that integration of the TA system into Tn6231 “such that all the transposon genes shared a single promoter region” permits coordinated TA and TnpR expression and may facilitate temporary inhibition of cell division while resolving the multimers, promoting plasmid persistence. In this light, it is interesting that the ccd TA system of Escherichia coli plasmid F is in an operon with a resolvase-encoding gene [319][320].
Expression of the TA module from the tnpA/tnpR promoter at the time of the transposition burst could transiently increase invasion efficiency (“addiction”) over and above that provided by the endogenous TA regulation system. If the transposon is on a molecule (e.g. a conjugative plasmid) that is unable to replicate vegetatively in the new host, expression of the TA module without transposition to a stable replicon would lead to loss of the transposon and consequent cell death, whereas cells in which transposition had occurred would survive and give rise to a new population in which all cells would contain the Tn. This might be seen as a “take me or die” mechanism [34], a notion which could be explored experimentally.
Conclusion and Future.
The Tn3 family is widely spread and diverse as we have underlined and illustrated here. There is some understanding of the different evolutionary pathways and mechanisms which have permitted family members to sequester a large set of passenger genes widely variable functions and to shuffle them between and within both plasmids and chromosomes. Although much is known about a number of model Tn3 family members, there remain a number of open questions. In particular, historically this has proved recalcitrant to analysis in spite of much effort from their discovery in the 1970s to the present day.
Recent studies with Tn4330 however may have unlocked a door to understanding Tn3 family transposition in molecular detail. The structural studies using cryo-em have provided precious information as to the location and function of a large number of domains in the exceptionally long transposases. The studies point to the way in which the transpososome may be assembled although additional analyses are essential to a full understanding of docking of target DNA and its place in the assembly pathway. In addition, it is at present unclear how duplication of this family occurs during transposition: how it may recruit replication enzymes, whether replication initiates from one particular end, or, indeed whether it involves parasitizing existing replication forks in the target. The phenomenon of immunity is also not understood although it is clear that, mutationally, it is linked to transposition activity. In the absence of an ATPase activity, it seems unlikely that it occurs with the same mechanism as does that of bacteriophage Mu or transposon Tn7.
Bibliography
- ↑ 1.0 1.1 Hedges RW, Jacob AE . Transposition of ampicillin resistance from RP4 to other replicons. - Mol Gen Genet: 1974, 132(1);31-40 [PubMed:4609125] [DOI]
- ↑ <pubmed>4609125</pubmed>
- ↑ Meynell E, Data N. Mutant Drug Resistant Factors of High transmissibility. Nature. 1967;214:885–887.
- ↑ 4.0 4.1 <pubmed>1093180</pubmed>
- ↑ 5.0 5.1 <pubmed>796463</pubmed>
- ↑ 6.0 6.1 6.2 <pubmed>606841</pubmed>
- ↑ 7.0 7.1 <pubmed>PMC232870</pubmed>
- ↑ 8.0 8.1 8.2 8.3 8.4 <pubmed>391406</pubmed>
- ↑ <pubmed>PMC319721</pubmed>
- ↑ 10.0 10.1 10.2 <pubmed>PMC218601</pubmed>
- ↑ 11.0 11.1 <pubmed>PMC383507</pubmed>
- ↑ 12.0 12.1 12.2 <pubmed>322280</pubmed>
- ↑ 13.0 13.1 <pubmed>PMC294055</pubmed>
- ↑ 14.0 14.1 <pubmed>340918</pubmed>
- ↑ 15.0 15.1 15.2 <pubmed>745233</pubmed>
- ↑ <pubmed>6297786</pubmed>
- ↑ 17.0 17.1 <pubmed>6271456</pubmed>
- ↑ <pubmed>PMC408333</pubmed>
- ↑ <pubmed>PMC246114</pubmed>
- ↑ Deonier RC, Yun K, Kuppermann M. Gamma delta-mediated deletions of chromosomal segments on F- prime plasmids. MolGenGenet. 1983;190:42–50.
- ↑ <pubmed>PMC413041</pubmed>
- ↑ <pubmed>2993789</pubmed>
- ↑ <pubmed>PMC294604</pubmed>
- ↑ 24.00 24.01 24.02 24.03 24.04 24.05 24.06 24.07 24.08 24.09 24.10 24.11 24.12 24.13 24.14 24.15 24.16 24.17 24.18 24.19 24.20 24.21 <pubmed>26350313</pubmed>
- ↑ 25.0 25.1 25.2 <pubmed>377024</pubmed>
- ↑ 26.0 26.1 26.2 26.3 26.4 26.5 <pubmed>PMC4337579</pubmed>
- ↑ <pubmed>PMC98933</pubmed>
- ↑ 28.0 28.1 28.2 28.3 28.4 28.5 28.6 28.7 28.8 <pubmed>22878084</pubmed>
- ↑ 29.0 29.1 29.2 29.3 <pubmed>2555940</pubmed>
- ↑ 30.0 30.1 30.2 30.3 <pubmed>2548736</pubmed>
- ↑ 31.0 31.1 31.2 31.3 31.4 <pubmed>PMC458404</pubmed>
- ↑ 32.0 32.1 32.2 32.3 <pubmed>PMC135285</pubmed>
- ↑ <pubmed>PMC1347377</pubmed>
- ↑ 34.0 34.1 34.2 34.3 34.4 34.5 34.6 34.7 34.8 34.9 <pubmed>PMC7157771</pubmed>
- ↑ 35.0 35.1 35.2 35.3 <pubmed>6312271</pubmed>
- ↑ 36.0 36.1 <pubmed>6294711</pubmed>
- ↑ <pubmed>1660177</pubmed>
- ↑ 38.0 38.1 <pubmed>3007931</pubmed>
- ↑ <pubmed>6327411</pubmed>
- ↑ <pubmed>PMC321896</pubmed>
- ↑ 41.0 41.1 41.2 <pubmed>6314094</pubmed>
- ↑ Heffron F. Tn3' ' and its relatives. In: James A. S, editor. Mobile' ' Genetic Elements. New York.: Academic Press,; 1983. p. 223–260.
- ↑ 43.0 43.1 43.2 43.3 43.4 43.5 43.6 43.7 <pubmed>1963947</pubmed>
- ↑ <pubmed>3005130</pubmed>
- ↑ 45.0 45.1 45.2 <pubmed>14553919</pubmed>
- ↑ <pubmed>PMC90453</pubmed>
- ↑ <pubmed>31624505</pubmed>
- ↑ <pubmed>31624505</pubmed>
- ↑ <pubmed>6298184</pubmed>
- ↑ <pubmed>2991421</pubmed>
- ↑ <pubmed>10477306</pubmed>
- ↑ <pubmed>10477306</pubmed>
- ↑ <pubmed>2991421</pubmed>
- ↑ <pubmed>2991421</pubmed>
- ↑ <pubmed>377024</pubmed>
- ↑ <pubmed>13727669</pubmed>
- ↑ <pubmed>340913</pubmed>
- ↑ 58.0 58.1 58.2 58.3 58.4 58.5 58.6 58.7 58.8 <pubmed>PMC103744</pubmed>
- ↑ 59.0 59.1 59.2 59.3 <pubmed>PMC221794</pubmed>
- ↑ <pubmed>PMC217832</pubmed>
- ↑ 61.0 61.1 61.2 <pubmed>2991421</pubmed>
- ↑ 62.0 62.1 <pubmed>PMC149298</pubmed>
- ↑ <pubmed>PMC2838034</pubmed>
- ↑ 64.0 64.1 64.2 <pubmed>2992807</pubmed>
- ↑ <pubmed>2560119</pubmed>
- ↑ <pubmed>PMC196970</pubmed>
- ↑ 67.0 67.1 67.2 <pubmed>PMC203974</pubmed>
- ↑ <pubmed>6260376</pubmed>
- ↑ <pubmed>377009</pubmed>
- ↑ <pubmed>PMC216121</pubmed>
- ↑ <pubmed>22142724</pubmed>
- ↑ <pubmed>2891921</pubmed>
- ↑ <pubmed>2968517</pubmed>
- ↑ <pubmed>PMC171412</pubmed>
- ↑ 75.0 75.1 <pubmed>PMC196104</pubmed>
- ↑ 76.0 76.1 76.2 <pubmed>PMC89148</pubmed>
- ↑ 77.0 77.1 <pubmed>PMC404624</pubmed>
- ↑ 78.0 78.1 <pubmed>PMC2258525</pubmed>
- ↑ <pubmed>PMC162963</pubmed>
- ↑ <pubmed>8380148</pubmed>
- ↑ <pubmed>10049255</pubmed>
- ↑ <pubmed>15131146</pubmed>
- ↑ <pubmed>18180362</pubmed>
- ↑ <pubmed>10548738</pubmed>
- ↑ <pubmed>10548738</pubmed>
- ↑ <pubmed>26802071</pubmed>
- ↑ <pubmed>11470367</pubmed>
- ↑ <pubmed>12007800</pubmed>
- ↑ <pubmed>PMC139670</pubmed>
- ↑ <pubmed>PMC127467</pubmed>
- ↑ <pubmed>PMC127047</pubmed>
- ↑ 92.0 92.1 <pubmed>15135523</pubmed>
- ↑ <pubmed>PMC182628</pubmed>
- ↑ 94.0 94.1 <pubmed>7830551</pubmed>
- ↑ <pubmed>PMC549287</pubmed>
- ↑ <pubmed>7830551</pubmed>
- ↑ <pubmed>23887414</pubmed>
- ↑ <pubmed>PMC2292522</pubmed>
- ↑ <pubmed>PMC2681555</pubmed>
- ↑ <pubmed>PMC3294963</pubmed>
- ↑ <pubmed>PMC3294926</pubmed>
- ↑ <pubmed>PMC2944623</pubmed>
- ↑ <pubmed>20660684</pubmed>
- ↑ <pubmed>20660684</pubmed>
- ↑ <pubmed>22733068</pubmed>
- ↑ <pubmed>11257029</pubmed>
- ↑ <pubmed>19400638</pubmed>
- ↑ 108.0 108.1 108.2 <pubmed>2830457</pubmed>
- ↑ <pubmed>PMC103765</pubmed>
- ↑ 110.0 110.1 <pubmed>PMC107244</pubmed>
- ↑ 111.0 111.1 111.2 111.3 111.4 111.5 111.6 <pubmed>15009901</pubmed>
- ↑ <pubmed>PMC95431</pubmed>
- ↑ <pubmed>19332822</pubmed>
- ↑ 114.0 114.1 114.2 114.3 <pubmed>PMC213150</pubmed>
- ↑ 115.0 115.1 115.2 115.3 <pubmed>PMC457184</pubmed>
- ↑ <pubmed>15491368</pubmed>
- ↑ 117.0 117.1 117.2 117.3 <pubmed>9274008</pubmed>
- ↑ 118.0 118.1 118.2 <pubmed>12427948</pubmed
- ↑ 119.0 119.1 <pubmed>12547188</pubmed>
- ↑ <pubmed>15856217</pubmed>
- ↑ 121.0 121.1 121.2 <pubmed>10822806</pubmed>
- ↑ <pubmed>15009901</pubmed>
- ↑ <pubmed>PMC52498</pubmed>
- ↑ <pubmed>12948488</pubmed>
- ↑ <pubmed>PMC1082539</pubmed>
- ↑ <pubmed>PMC1352228</pubmed>
- ↑ <pubmed>32871211</pubmed>
- ↑ Galas DJ, Chandler M. Bacterial Insertion Sequences. In: Berg DE, Howe MM, editors. Mob DNA. Washington, D.C.: American Society for Microbiology; 1989. p. 109–162.
- ↑ <pubmed>PMC7986276</pubmed>
- ↑ 130.0 130.1 <pubmed>23802695</pubmed>
- ↑ <pubmed>30207068</pubmed>
- ↑ 132.0 132.1 132.2 <pubmed>PMC201273</pubmed>
- ↑ <pubmed>11495991</pubmed>
- ↑ <pubmed>PMC111268</pubmed>
- ↑ <pubmed>12949179</pubmed>
- ↑ <pubmed>PMC523222</pubmed>
- ↑ 137.0 137.1 <pubmed>PMC127583</pubmed>
- ↑ <pubmed>PMC95461</pubmed>
- ↑ <pubmed>11976102</pubmed>
- ↑ <pubmed>PMC7039861</pubmed>
- ↑ <pubmed>PMC168534</pubmed>
- ↑ <pubmed>PMC4133298</pubmed>
- ↑ 143.0 143.1 <pubmed>PMC4588545</pubmed>
- ↑ <pubmed>PMC3751643</pubmed>
- ↑ <pubmed>PMC4579464</pubmed>
- ↑ <pubmed>PMC3826837</pubmed>
- ↑ <pubmed>25691597</pubmed>
- ↑ <pubmed>19933107</pubmed>
- ↑ <pubmed>22781676</pubmed>
- ↑ <pubmed>21179091</pubmed>
- ↑ <pubmed>19933107</pubmed>
- ↑ <pubmed>22223738</pubmed>
- ↑ <pubmed>21106758</pubmed>
- ↑ <pubmed>22828628</pubmed>
- ↑ 155.0 155.1 <pubmed>6098802</pubmed>
- ↑ <pubmed>11376944</pubmed>
- ↑ 157.0 157.1 157.2 157.3 <pubmed>9159519</pubmed>
- ↑ <pubmed>17408691</pubmed>
- ↑ <pubmed>8387603</pubmed>
- ↑ Jobling MG, Peters SE, Ritchie DA. Plasmid-borne mercury resistance in aquatic bacteria. FEMS Microbiol Lett. 1988;49:31–37.
- ↑ <pubmed>6394954</pubmed>
- ↑ <pubmed>PMC183436</pubmed>
- ↑ <pubmed>9167257</pubmed>
- ↑ <pubmed>6316165</pubmed>
- ↑ <pubmed>11763242</pubmed>
- ↑ <pubmed>PMC210155</pubmed>
- ↑ <pubmed>10079080</pubmed>
- ↑ 168.0 168.1 <pubmed>1917975</pubmed>
- ↑ <pubmed>PMC168397</pubmed>
- ↑ <pubmed>19265693</pubmed>
- ↑ 171.0 171.1 <pubmed>8063107</pubmed>
- ↑ <pubmed>PMC304818</pubmed>
- ↑ <pubmed>3542022</pubmed>
- ↑ 174.0 174.1 174.2 174.3 174.4 174.5 174.6 <pubmed>16629665</pubmed>
- ↑ <pubmed>PMC5142620</pubmed>
- ↑ 176.0 176.1 <pubmed>385227</pubmed>
- ↑ <pubmed>PMC294047</pubmed>
- ↑ <pubmed>6294460</pubmed>
- ↑ <pubmed>6329911</pubmed>
- ↑ <pubmed>6308391</pubmed>
- ↑ 181.0 181.1 <pubmed>6090862</pubmed>
- ↑ <pubmed>6262296</pubmed>
- ↑ <pubmed>2986006</pubmed>
- ↑ <pubmed>PMC299513</pubmed>
- ↑ <pubmed>2843651</pubmed>
- ↑ <pubmed>2155858</pubmed>
- ↑ 187.0 187.1 <pubmed>1849179</pubmed>
- ↑ <pubmed>2162965</pubmed>
- ↑ <pubmed>7744253</pubmed>
- ↑ <pubmed>PMC553657</pubmed>
- ↑ <pubmed>2821273</pubmed>
- ↑ <pubmed>9630232</pubmed>
- ↑ 193.0 193.1 193.2 193.3 <pubmed>PMC5293067</pubmed>
- ↑ <pubmed>8382339</pubmed>
- ↑ 195.0 195.1 195.2 195.3 <pubmed>8080658</pubmed>
- ↑ 196.0 196.1 196.2 196.3 <pubmed>22624153</pubmed>
- ↑ <pubmed>9097724</pubmed>
- ↑ 198.0 198.1 <pubmed>PMC3107681</pubmed>
- ↑ 199.0 199.1 <pubmed>10884228</pubmed>
- ↑ <pubmed>PMC7780238</pubmed>
- ↑ <pubmed>2172235</pubmed>
- ↑ <pubmed>9077464</pubmed>
- ↑ <pubmed>PMC1170910</pubmed>
- ↑ <pubmed>8382339</pubmed>
- ↑ <pubmed>8080658</pubmed>
- ↑ <pubmed>1652540</pubmed>
- ↑ Oger C. End-synapsis and target integration during replicative transpositon of the Tn3-family transposon Tn4430 [Doctoral dissertation]. Université de Louvain la Neuve; 2018.
- ↑ <pubmed>PMC3977044</pubmed>
- ↑ <pubmed>PMC7292550</pubmed>
- ↑ 210.0 210.1 210.2 Shkumatov AV, Aryanpour N, Oger CA, Goossens G, Hallet BF, Efremov RG. Metamorphism of catalytic domain controls transposition in Tn3 family transposases. BioRxiv. 2022 Feb 24.
- ↑ Hallet B. Tn4430 AFM.
- ↑ <pubmed>22624153</pubmed>
- ↑ 213.0 213.1 <pubmed>28096365</pubmed>
- ↑ <pubmed>PMC234940</pubmed>
- ↑ 215.0 215.1 215.2 215.3 215.4 <pubmed>6278249</pubmed>
- ↑ <pubmed>672895</pubmed>
- ↑ <pubmed>6244100</pubmed>
- ↑ <pubmed>6317201</pubmed>
- ↑ <pubmed>PMC282071</pubmed>
- ↑ <pubmed>2544738</pubmed>
- ↑ <pubmed>PMC3703974</pubmed>
- ↑ <pubmed>PMC1170876</pubmed>
- ↑ <pubmed>PMC312388</pubmed>
- ↑ <pubmed>26104363</pubmed>
- ↑ 225.0 225.1 <pubmed>6271635</pubmed>
- ↑ 226.0 226.1 <pubmed>PMC390066</pubmed>
- ↑ 227.0 227.1 227.2 227.3 <pubmed>3009272</pubmed>
- ↑ <pubmed>6278250</pubmed>
- ↑ <pubmed>PMC209839</pubmed>
- ↑ 230.0 230.1 <pubmed>PMC213758</pubmed>
- ↑ <pubmed>PMC213151</pubmed>
- ↑ <pubmed>6281440</pubmed>
- ↑ <pubmed>PMC2916420</pubmed>
- ↑ <pubmed>9077463</pubmed>
- ↑ <pubmed>6092854</pubmed>
- ↑ <pubmed>6092853</pubmed>
- ↑ <pubmed>PMC341238</pubmed>
- ↑ <pubmed>PMC210914</pubmed>
- ↑ <pubmed>2168370</pubmed>
- ↑ <pubmed>2168370</pubmed>
- ↑ <pubmed>392228</pubmed>
- ↑ 242.0 242.1 242.2 <pubmed>6313485</pubmed>
- ↑ 243.0 243.1 243.2 243.3 243.4 243.5 <pubmed>11994145</pubmed>
- ↑ <pubmed>PMC3775659</pubmed>
- ↑ 245.0 245.1 <pubmed>PMC4384473</pubmed>
- ↑ 246.0 246.1 246.2 246.3 246.4 <pubmed>6269756</pubmed>
- ↑ 247.0 247.1 247.2 247.3 247.4 <pubmed>6290077</pubmed>
- ↑ 248.0 248.1 <pubmed>PMC319581</pubmed>
- ↑ 249.0 249.1 <pubmed>PMC553467</pubmed>
- ↑ 250.0 250.1 250.2 250.3 <pubmed>PMC555234</pubmed>
- ↑ <pubmed>26104558</pubmed>
- ↑ <pubmed>6094833</pubmed>
- ↑ <pubmed>PMC457183</pubmed>
- ↑ 254.0 254.1 254.2 <pubmed>2850169</pubmed>
- ↑ 255.0 255.1 <pubmed>PMC345424</pubmed>
- ↑ <pubmed>PMC551726</pubmed>
- ↑ 257.0 257.1 <pubmed>PMC306530</pubmed>
- ↑ 258.0 258.1 <pubmed>6301692</pubmed>
- ↑ <pubmed>6313351</pubmed>
- ↑ <pubmed>2998784</pubmed>
- ↑ <pubmed>PMC397197</pubmed>
- ↑ <pubmed>2990045</pubmed>
- ↑ <pubmed>15994378</pubmed>
- ↑ <pubmed>PMC1483221</pubmed>
- ↑ 265.0 265.1 265.2 <pubmed>PMC2428073</pubmed>
- ↑ <pubmed>33405333</pubmed>
- ↑ Montaño SP, Rowland S-J, Fuller JR, Burke ME, MacDonald AI, Boocock MR, Stark WM, Rice PA. Structural basis for topological regulation of Tn3 resolvase. BioRxiv. 2021 Dec 8.
- ↑ 268.0 268.1 <pubmed>PMC345658</pubmed>
- ↑ <pubmed>2993811</pubmed>
- ↑ <pubmed>PMC554474</pubmed>
- ↑ <pubmed>6318048</pubmed>
- ↑ <pubmed>2993810</pubmed>
- ↑ <pubmed>PMC310539</pubmed>
- ↑ 274.0 274.1 <pubmed>2175363</pubmed>
- ↑ <pubmed>2175362</pubmed>
- ↑ <pubmed>1327971</pubmed>
- ↑ 277.0 277.1 277.2 <pubmed>9663657</pubmed>
- ↑ <pubmed>3524854</pubmed>
- ↑ <pubmed>PMC413488</pubmed>
- ↑ <pubmed>11972771</pubmed>
- ↑ 281.0 281.1 <pubmed>15813731</pubmed>
- ↑ <pubmed>16553879</pubmed>
- ↑ Structural basis for topological regulation of Tn3 resolvase. Sherwin P. Montano, Sally-J. Rowland, James R. Fuller, Mary E. Burke, Alasdair I. MacDonald, Martin R. Boocock, W. Marshall Stark, Phoebe A. Rice. bioRxiv 2021.12.07.471667; doi: https://doi.org/10.1101/2021.12.07.471667
- ↑ <pubmed>PMC401285</pubmed>
- ↑ <pubmed>PMC6007550</pubmed>
- ↑ <pubmed>3018445</pubmed>
- ↑ 287.0 287.1 287.2 287.3 <pubmed>PMC205437</pubmed>
- ↑ 288.0 288.1 288.2 288.3 <pubmed>7608077</pubmed>
- ↑ 289.0 289.1 <pubmed>26104563</pubmed>
- ↑ <pubmed>PMC1170750</pubmed>
- ↑ <pubmed>11340053</pubmed>
- ↑ <pubmed>9288963</pubmed>
- ↑ <pubmed>PMC450016</pubmed>
- ↑ <pubmed>PMC2859247</pubmed>
- ↑ <pubmed>18439894</pubmed>
- ↑ Structural basis for topological regulation of Tn3 resolvase. Sherwin P. Montano, Sally-J. Rowland, James R. Fuller, Mary E. Burke, Alasdair I. MacDonald, Martin R. Boocock, W. Marshall Stark, Phoebe A. Rice. bioRxiv 2021.12.07.471667; doi: https://doi.org/10.1101/2021.12.07.471667
- ↑ <pubmed>PMC2847244</pubmed>
- ↑ <pubmed>PMC209757</pubmed>
- ↑ <pubmed>12169600</pubmed>
- ↑ <pubmed>2851712</pubmed>
- ↑ <pubmed>26104463</pubmed>
- ↑ <pubmed>12169600</pubmed>
- ↑ <pubmed>16122561</pubmed>
- ↑ 304.0 304.1 304.2 304.3 304.4 <pubmed>PMC4840908</pubmed>
- ↑ <pubmed>PMC219208</pubmed>
- ↑ <pubmed>PMC323463</pubmed>
- ↑ 307.0 307.1 307.2 <pubmed>PMC3141249</pubmed>
- ↑ 308.0 308.1 308.2 308.3 308.4 308.5 308.6 <pubmed>21819231</pubmed>
- ↑ 309.0 309.1 <pubmed>34975154</pubmed>
- ↑ <pubmed>PMC3710099</pubmed>
- ↑ <pubmed>12010492</pubmed>
- ↑ 312.0 312.1 <pubmed>PMC103759</pubmed>
- ↑ <pubmed>27159580</pubmed>
- ↑ <pubmed>20603017</pubmed>
- ↑ <pubmed>13373067</pubmed>
- ↑ <pubmed>14482966</pubmed>
- ↑ <pubmed>4598795</pubmed>
- ↑ <pubmed>PMC1202851</pubmed>
- ↑ <pubmed>PMC341330</pubmed>
- ↑ <pubmed>2511422</pubmed>