General Information/Transposase expression and activity
Contents
- 1 Transposase expression and activity
- 1.1 Impinging transcription
- 1.2 Sequestration of translation initiation signals
- 1.3 Programmed Translational Frameshifting
- 1.4 Programmed Transcriptional Frameshifting
- 1.5 Recoding suppression of stop codons using the unusual amino acids Pyrrolysine and Selenocysteine
- 1.6 Translation termination
- 1.7 Transposase stability
- 1.8 Co-translational binding and multimerization
- 1.9 Cleavage in Trans: A Committed Complex
- 1.10 Host factors
- 1.11 Over-production inhibition
- 2 Bibliography
Transposase expression and activity
While many of the classical mechanisms of controlling gene expression, such as the production of transcriptional repressors (IS1: [1][2]; IS2: [3] or translational inhibitors (anti-sense RNA in the case of IS10; see [4] are known to operate in Tpase expression, several other mechanisms have also been uncovered.
Impinging transcription
Many ISs have evolved mechanisms which attenuate their activation by impinging transcription following insertion into active host genes. This was originally observed in the case of IS1[5][6], for bacteriophage Mu[7] and IS50. To our knowledge other elements have not been examined.
This effect may be the result of disrupting complexes between Tpase and cognate DNA ends and could reflect either inhibition of transposase binding or disruption of extant transposase-end complexes.
In the case of bacteriophage Mu, transcription originating from within the element and impinging on the left end has also been shown to reduce activity[7]. It is possible that transcription disrupts the formation of intermediates including transposase and one or both Mu ends which lead to stable transposition complexes.
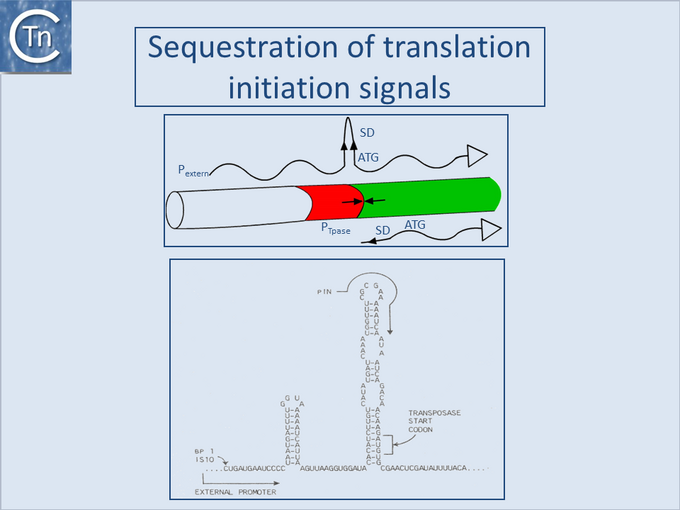
Sequestration of translation initiation signals
One such mechanism observed with IS10 and IS50, and potentially present in several other ISs, is the sequestering of translation initiation signals in an RNA secondary structure (Fig.21.1). These ISs carry inverted repeat sequences located close to the left end which include the ribosome binding site or translation initiation codon for the Tpase gene. Transcripts from the resident Tpase promoter include only the distal repeat unit which is unable to form the secondary structure, while transcripts from neighboring DNA include both repeats and would generate secondary structures in the mRNA which would sequester translation initiation signals[8][9]. This has been demonstrated experimentally for IS10 and IS50 but several additional insertion sequences carry such potential structures and might be expected to exhibit a similar mechanism (see [10]).
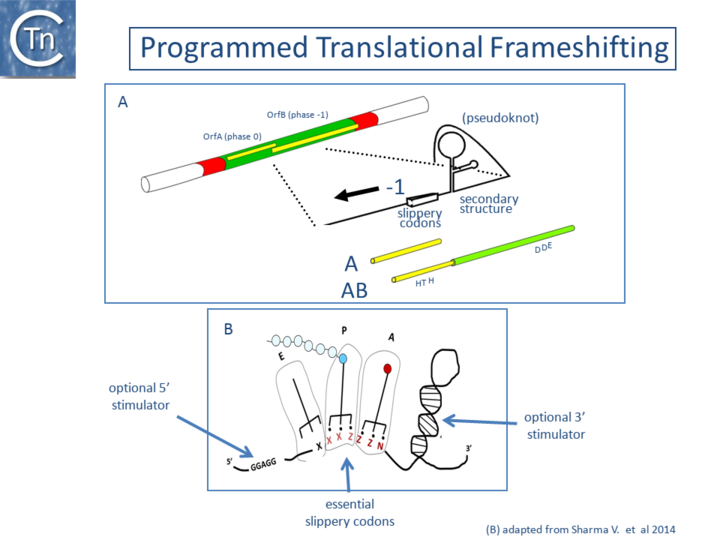
Programmed Translational Frameshifting
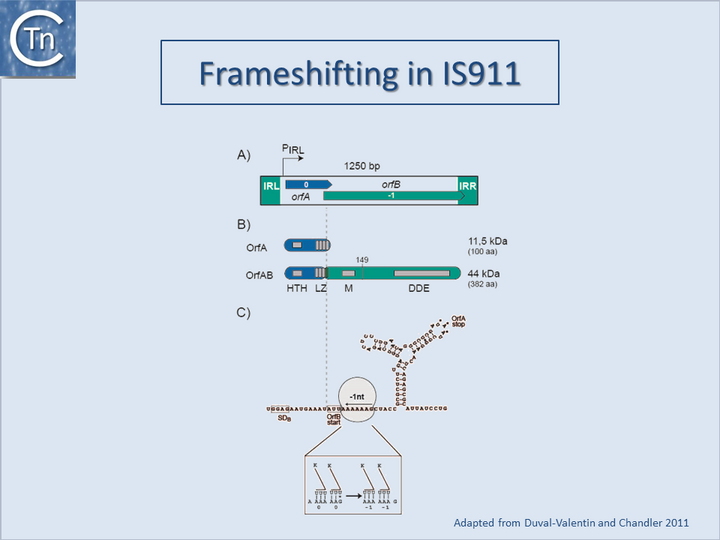
A second mechanism acts at the level of translation elongation and involves programmed translational frameshifting between two consecutive open reading frames (Fig.21.1). Typically a -1 frameshift is observed in which the translating ribosome slides one base upstream and resumes in the alternative phase. This generally occurs at the position of so-called slippery codons in a heptanucleotide sequence of the type X XXZ ZZN in phase 0 (where the bases paired with the anticodon are shown as triplets) which is read as XXX ZZZ N in the shifted -1 phase (Fig.21.2) (see e.g. [11][12][13], http://recode.genetics.utah.edu/). The sequence A AAA AAG is a common example of this type of heptanucleotide. Ribosomal shifting of this type is stimulated by structures in the mRNA which tend to impede the progression of the ribosome such as potential ribosome binding sites upstream or secondary structures (stem-loop structures and pseudoknots) downstream of the slippery codons. Translational control of transposition by frameshifting has been demonstrated both for IS1[14][2], and for members of the IS3 family (Fig.21.3) ([15]; see also [11]) but may also occur in several other IS elements (see for example IS5 and IS630 families). For IS1 and members of the IS3 family, the upstream frame appears to carry a DNA recognition domain whereas the downstream frame encodes the catalytic site. While the product of the upstream frame alone acts as a modulator of activity, presumably by binding to the IR sequences, frameshifting assembles both domains into a single protein, the Tpase, which directs the cleavages and strand transfer necessary for mobility of the element. The frameshifting frequency is thus critical in determining overall transposition activity. Although it has yet to be explored in detail, frameshifting could be influenced by host physiology thus coupling transposition activity to the state of the host cell.
Programmed Transcriptional Frameshifting
Another type of frameshifting may also occur in bacterial insertion sequences. This occurs at the transcriptional level and involves misreading and slippage of RNA polymerase on stretches of A residues. Although no real data is at present available, it may occur relatively frequently [16][17][18].
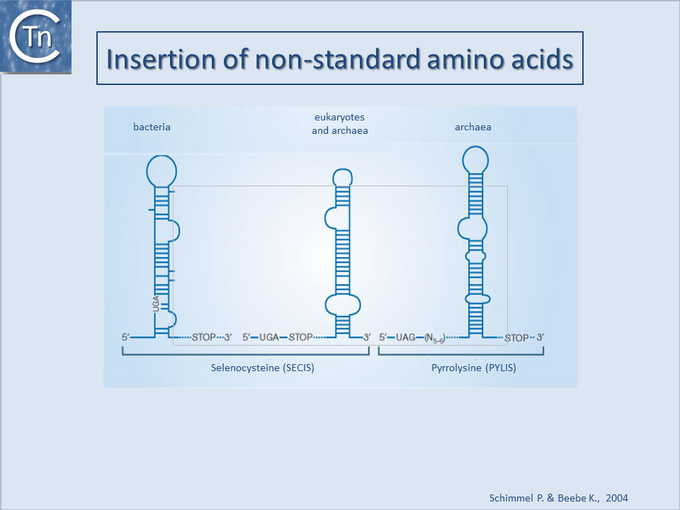
Recoding suppression of stop codons using the unusual amino acids Pyrrolysine and Selenocysteine
Another type of recoding which appears to occur in Methanosarcina, is the “suppression” of the stop codon, UAG, by insertion of Pyrrolysine. This was first noted in the Methylamine methyltransferases which are important in the production of methane by archaeal methanogens [19] identified an in-frame amber codon (TAG) in the trimethylamine methyltransferase genes of both M. barkeri and M. thermophila. However, at least in the case of M. barkeri, abundant quantities of the full-length protein could be obtained and it appeared that the TAG codon was read as Lys. This later proved to be the unusual amino acid pyrrolysine. Several IS copies in these archaeal methanogens carry TAG codons which are presumably “suppressed” by decoding as pyrrolysine.
In this framework, other stop codons are known to be suppressed by decoding as selenocysteine [20][21][22][23]. To our knowledge a single IS from the IS3 family, ISDvu3 from Desulfovibrio vulgaris, includes a selenocysteine inserted at a stop codon in its orfB frame (M. Land personal communication). Undoubtedly additional examples will be identified in the future.
The inclusion of these amino acids involves the presence of specific types of secondary structures in the mRNA (Fig.21.4).
Translation termination
A third potential mechanism derives from the observation that the translation termination codon of Tpase genes of certain elements is located within their IR sequences. Although to our knowledge, no extensive analysis of the significance of this arrangement has yet been undertaken, it seems possible that it may in some manner couple translation termination, transposase binding, and transposition activity. The transposase gene of several elements does not possess a termination codon. These include IS240C, a member of the IS6 family (Y. Chen and J.Mahillon unpublished), two members of the IS5 family, IS427[24] and ISMk1[25] and various members of the IS630 family including IS870 and ISRf1[26]. Instead, some of these elements insert into a relatively specific target sequence in which the target DR produced on insertion itself generates the Tpase termination codon (see: IS630 family). The relevance of this as a control mechanism has yet to be explored.
Transposase stability
Transposase stability can also contribute to the control of transposition activity. The Tpase of IS903 is sensitive to the E. coli Lon protease[27]. This sensitivity limits the activity of the Tpase both temporally and spatially and may provide an explanation for the observation that several Tpases function preferentially in cis. Indeed mutant IS903 Tpase derivatives have been isolated which exhibits an increased capacity to function in trans. These are more refractory to Lon degradation than the wildtype protein[28]. Some evidence that Lon may also be involved in regulating Tn5 (IS50) transposition has also been presented[9].
An observation which might also reflect Tpase instability is the temperature sensitive nature of IS1-mediated adjacent deletions in vivo[29], of Tn3 transposition[30] and of IS911 intramolecular recombination both in vivo and in vitro[31]. For IS911, incubation of the Tpase at 42°C results in an irreversible loss in activity. Further analyses showed that there was an increase in the proportion of transposase fragments some of which can presumably interact with full length transposase to inhibit its activity. This effect can be somewhat reduced by a series of mutation in the transposase gene whose function in stabilizing the transposase is as yet unknown[32].
Co-translational binding and multimerization
Certain prokaryotic IS transposases show a strong preference for acting on the element from which they are expressed rather than on other copies of the same element in the cell. This phenomenon of “cis” preference presumably serves to prevent general activation of several identical IS copies by any “accidental” (stochastic) transposase expression from a single IS. Several different IS such as IS1[5], IS10[33], IS50[34], IS903[35] and IS911[36] (see [37][27] and references therein) exhibit this regulatory phenotype but “cis” preference may be the result of a combination of diverse mechanisms. Thus the Lon protease enhances “cis” preference of the IS903 transposase [27]. Transposition is enhanced in the absence of Lon and can be overcome by increased transposase expression[28]. For IS10, it is influenced by translation levels, Tpase mRNA half-life and translation efficiency[38][39].
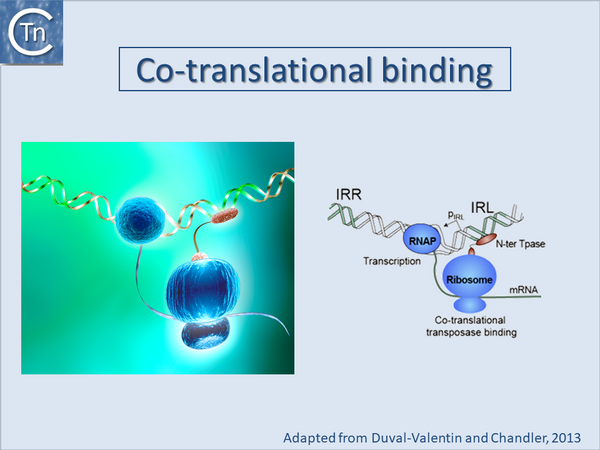
Another mechanism, co-translational binding based on tight coupling between prokaryotic transcription and translation, was proposed to explain the inability to complement a Tpase mutant of IS903[35] and, more specifically, for Tn5[40].
Some full length IS transposases bind weakly to their cognate IR but the isolated DNA binding domain can bind more strongly. This has been observed for transposases of several elements including IS1[41] and IS30[42][43] and has also been observed for that of IS911. Early studies using band shift assays demonstrated that full length OrfAB binds the IRs only weakly and that OrfA binding was even lower or undetectable[44][45]. However, a truncated version of OrfAB, OrfAB [1-149], which is amputated for the C-terminal catalytic domain bound both ends avidly[46] (see also "Transposase Stability"). It is important to note this implies that, in many in vitro systems, the majority of transposase is thus likely to be inactive or only partially active since it would not bind stably to its substrate. The observations suggest that the C-terminal (C-ter) domain inhibits specific binding by the sequence-specific N-terminal DNA binding domain possibly by steric masking (Fig.21.5). This idea is consistent with the observation that IS10 transposase activity is increased by partial denaturation (for example by treatment with low alcohol concentrations; [47]). It is also consistent with the observation that the OrfAB protein of IS2 can bind the IS2 IRs when it carries a large GFP tag[48][49].
One biological explanation for cis preference is that the nascent N-ter domain might fold before completion of translation of the C-ter domain and the nascent protein could initiate binding directly to the closest IS end. Once bound, it would no longer be sensitive to masking by the C-ter domain. If binding fails to occur after translation of the N-ter DNA binding domain, continuing translation and folding of the C-ter domain would then mask the DNA binding domain resulting in an inactive protein. This implies that binding necessary for subsequent catalysis would occur only transitorily early in translation (Fig.21.5).
Direct evidence for co-translational binding was provided for IS911 using an in vitro transcription/translation system[36] where it was also demonstrated that reducing the efficiency of the -1 translational frameshifting required for IS911 transposase expression resulted in an increase in binding of a nascent transposase peptide. This is presumably because slowing the frameshifting process increases the time that the N-terminal part of the protein (which carries the sequence-specific DNA binding domain) is present on the ribosome enhancing its probability of binding to a neighboring IS end. It is interesting to note that in many IS, the DNA binding domain which recognizes the IR is located at the N-terminal end of the protein which is translated first.
One of the remaining questions concerns transposase multimerization. They must form multimers within the transpososome at some stage in the transposition pathway. Some transposases are monomeric in the absence of DNA (e.g. MuA and Tn5; [50][51][52]) while others are multimeric dimeric in solution (e.g. INHIV-1; [53]; [54]; [55]; [56]).
In view of the possibility that many transposases undergo co-translational binding, and the observation that several different purified full-length transposases bind poorly to the ends of their cognate transposon (while the isolated N-terminal DNA binding domain alone binds robustly), it must be emphasized that purified transposases are probably largely inactive. This must be taken into account when assessing transposase properties.
A recent study has provided support for the idea that transposases may also be able to multimerize cotranslationally. This study, has shown that bacterial luciferase subunits LuxA and LuxB may assemble cotranslationally in vivo. This process requires ribosome-associated trigger factor. This chaperone apparently delays subunit interactions until the LuxB dimer interface is available[57].
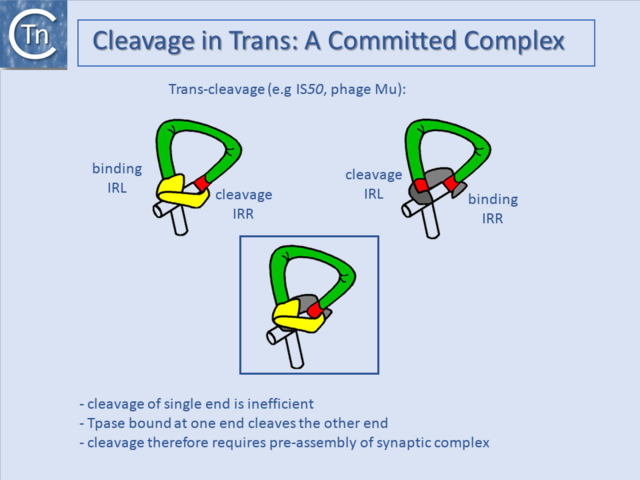
Cleavage in Trans: A Committed Complex
Another level of regulation often occurs within the transpososome (see "Reaction Mechanisms") itself. Transposases bound to their specific recognition sequence, generally at both ends of the TE, form a Synaptic (SC) or Paired-End Complex (PEC) with a precise architecture for activity, and the transposase bound at one end is used to cleave the opposite end (Fig.21.6) (e.g. [58] and [59]). This ensures that the correct SC is formed prior to cleavage and strand transfer and thus avoids the generation of nonproductive cleavage products, which would cause damage to the donor DNA molecule.
Host factors
Transposition activity is frequently modulated by various host factors. These effects are generally specific for each element. A non-exhaustive list of such factors includes the DNA chaperones (or histone-like proteins), IHF, HU, HNS, and FIS, the replication initiator DnaA, the protein chaperone/proteases ClpX, ClpP, and ClpA, the SOS control protein LexA, and the Dam DNA methylase. In addition, proteins which govern DNA supercoiling in the cell can also influence transposition.
IHF, HU, HNS, and FIS
DNA chaperones may play roles in assuring the correct three dimensional architecture in the evolution of various nucleoprotein complexes necessary for productive transposition. They may also be involved in regulating Tpase expression. IHF, HU, HNS, and FIS have all been variously implicated in the case of bacteriophage Mu, in the control of Mu gene expression or directly in the transposition process (see [60] for review).
Several elements carry specific binding sites for IHF within, or close to, their terminal IRs. These can lie within (e.g. IS1: [61]; IS903: see [62] or close to (IS10 [4]) the Tpase promoter. IHF appears to influence the nature of IS10 transposition products by binding to a site 43 bp from one end [63][64][65].
IHF also stimulates Tpase binding to the ends of the Tn3 family member, Tn1000 or γδ [66]. Ironically, although IS1 was the first element in which IHF sites were identified (one within each IR), conditions have not yet been found in which IHF shows a clear effect on transposition or gene expression (D. Zerbib and M. Chandler, unpublished results).
The multiple roles of several of these proteins in both the IS10 and Tn5 (IS50) systems and the dynamics of their involvement has been determined in detail[64][67][68][69][70][71][72][73].
The histone-like nucleoid structuring protein H-NS, a global transcriptional regulator, has also been implicated in the regulation of bacterial transposition systems, including Tn10[70][72][73]. It appears to promote transposition by binding directly to the transposition complex (or transpososome).
DnaA
In the case of IS50, an element of the same family as IS10, both the protein Fis and the replication initiator protein DnaA have been reported to intervene in transposition (see [74]). Finally another "histone-like" protein, HNS, has been reported to stimulate transposition of IS1 in certain circumstances[75].
Although their mode of action is at present unknown, several other host proteins with otherwise entirely different functions have been implicated in transposition.
Accessory proteins: Acyl carrier protein (ACP), ribosomal protein L29, PepA and ArgR Acyl carrier protein (ACP) was independently shown to stimulate 3' end cleavage of Tn3 by its cognate Tpase[76] and, together with ribosomal protein L29, to greatly increase binding of TnsD (a protein involved in Tn7 target selection) to the chromosomal insertion site, attTn7 [72]. Moreover ACP and L29 moderately stimulate Tn7 transposition in vitro while L29 alone has a significant stimulatory effect in vivo [72]. The mode of action of these proteins may be similar to that of the accessory proteins PepA and ArgR which modify the architecture of the synaptic complex in certain XerC/XerD-mediated site-specific recombination reactions[77].
Accessory proteins: Acyl carrier protein (ACP), ribosomal protein L29, PepA and ArgR
Acyl carrier protein (ACP) was independently shown to stimulate 3' end cleavage of Tn3 by its cognate Tpase [76] and, together with ribosomal protein L29, to greatly increase binding of TnsD (a protein involved in Tn7 target selection) to the chromosomal insertion site, attTn7 [78]. Moreover ACP and L29 moderately stimulate Tn7 transposition in vitro while L29 alone has a significant stimulatory effect in vivo [78]. The mode of action of these proteins may be similar to that of the accessory proteins PepA and ArgR which modify the architecture of the synaptic complex in certain XerC/XerD-mediated site-specific recombination reactions [77].
ClpX, ClpP, and Lon
Certain factors involved in protein "management" such as ClpX, ClpP, and Lon have been implicated in transposition. ClpX is essential for bacteriophage Mu growth[79] where it is required for disassembling the transposase-DNA complex or the transpososome strand transfer complex in preparation for the assembly of a replication complex[80][81]. Recognition of Mu transposase, pA, by ClpX requires the terminal 10 amino acids of pA[82]. Together with ClpP, ClpX also plays a role in proteolysis of the Mu repressor[83][84]. The Lon protease is implicated in proteolysis of the IS903 transposase [27].
At present the involvement of these proteins in the transposition of other elements has not been well documented.
SOS system, RecA, RecBC
The third class of host factor includes host cell systems which act to limit DNA damage and maintain chromosome integrity. Studies with IS10 (see [4]) and IS1[85] have demonstrated that high levels of Tpase in the presence of suitable terminal IRs lead to induction of the host SOS system. As discussed previously[86], some controversy still exists concerning the role of RecA in Tn5 (IS50) transposition[87][88][89]. Reznikoff and colleagues have provided genetic evidence that transposition is inhibited by induction of the SOS system in a manner which does not require the proteolytic activity of RecA[89]. On the other hand, Tessman and collaborators[87][88][90] using a different transposition assay have found that constitutive SOS conditions actually enhance Tn5 transposition. Moreover, using yet another assay system, Ahmed[91] has concluded that intermolecular transposition of Tn5 is stimulated by RecA. Further investigation is clearly required to understand these apparently incompatible results.
Ahmed has also concluded that intermolecular transposition of the IS1-based transposon, Tn9, behaves in a similar way to that of Tn5 with respect to the recA allele[91]. In contrast, however, the frequency of adjacent deletions mediated by IS1 was significantly increased in the absence of RecA. This has received some independent support using a physical assay where it was shown that deletion products accumulate in a recA but not in a wildtype host. Moreover, like IS1 induction of the SOS system, accumulation of such adjacent deletions was dependent on recBC (Zablweska et al., unpublished observations). The recBC genes are also implicated in the behavior of transposons such as Tn10 and Tn5[91] where they affect precise and imprecise excision in a process independent of transposition per se. This is more pronounced with composite transposons in which the component insertion sequences IS10 and IS50 are present as inverted repeats, and is stimulated when the transposon is carried by a transfer-proficient conjugative plasmid. It seems probable that such excisions occur by a process involving replication fork slippage (see [92][93] for further discussion).
PolI and gyrase
Both DNA polymerase I [94][95][96] and DNA gyrase[97][98] are implicated in the transposition of Tn5. While the effect of gyrase may reflect a requirement for optimal levels of supercoiling, the role of PolI remains a matter of speculation. It may be involved in DNA synthesis necessary to repair the single strand gaps resulting from staggered cleavage of the target and which gives rise to the DRs. DNA gyrase has also been shown to be important in transposition of bacteriophage Mu[99][100].
Dam methylase
Another host function, the Dam DNA methylase can be important in modulating both Tpase expression and activity. IS10, IS50 and IS903 all carry methylation sites (GATC) in the transposase promoter regions and in each case, promoter activity is increased in a dam-host[101][102]. Additional evidence has been presented that the methylation status of GATC sites within the terminal inverted repeats also modulates the activity of these ends[101]. For IS50, this can now be understood in terms of steric interference in the transposase active site, as recently revealed by the determination of the crystal structure of a synpatic complex including its Tpase and a pair of precleaved transposon ends[103]. Similar methylation sites have been previously observed in IS3, IS4, and IS5. A survey of the elements included in the data base has shown that most groups or families contain members which have GATC sites within the first 50 bp of one or both extremities. The IS3, IS5 and IS256 families include the most members carrying such sites. Except for IS3 itself where strong stimulation of transposition has been observed in a dam-host, in most of these cases the biological relevance of these sites is unknown. Moreover, it should be pointed out that the probability that any 100 bp DNA sequence carries the GATC tetranucleotide is about 40%. The role of Dam methylation in IS10 and IS50 transposition is described in detail in the appropriate sections dealing with these elements.
Metabolic control elements
In a screen of over 20,000 independent insertion mutants for host factors that influence IS903 transposition the Derbyshire lab isolated more than 100 mutants that increased or decreased transposition and also altered its timing during colony growth[104]. These included independent mutations in a gene required for fermentative metabolism during anaerobic growth resulting in “early” transposition during colony growth and was suppressed by addition of fumarate, and other mutations in genes associated with DNA metabolism, intermediary metabolism, transport, cellular redox, protein folding and proteolysis. Other mutations were isolated in pur genes involved in purine biosynthesis. Further analysis suggested that this phenotype was due to a requirement for GTP in IS903 transposition[105]. It should be noted that some of these mutants also affected transposition of IS10 and of Tn552.
Hfq
Finally, the RNA chaperone Hfq has also been implicated in the regulation of Tn10 transposition by promoting RNAout interaction with transposase mRNA[106][107][108]. There is also some suggestion that SmAP1, the archaeal Hfq homolog, may also play a role in regulation of transposition of several different archaeal IS [109] but this remains to be tested directly.
Over-production inhibition
Certain transposons appear to be subject to a mode of regulation known as over-expression inhibition. This was first observed with the eukaryotic transposons Tc1/mariner Lampe[110][111][112] where increasing the concentration of transposase results in a reduction in the level of transposition. It was subsequently observed with the sleeping beauty transposon[113][114]. It also occurs in vivo in mice[115][116].
The biological rational for this is that “infection” of a naïve cell by the transposon results in a burst of transposition which is then attenuated by overproduction inhibition. This is then followed by gradual decay of the transposon.
The Chalmers lab[117] has provided an interesting and compelling explanation of this effect. Using the mariner family transposon Hsmar1 they present convincing data implying that overproduction inhibition occurs during transpososome assembly and is due to a combination of the multimeric state of the transposase coupled with competition for transposase binding sites at the Hsmar1 ends[118][119]. The model (assembly-site-occlusion model) is based on the presence of transposase multimers (dimers) to the exclusion of monomers – in other words, end-binding required a dimeric transposase. At low transposase/transposon ratios, one dimer can bind both transposon ends resulting in the ordered assembly of the transpososome. An increase in the transposase dimer/transposon ratio results in binding of dimers to both transposon ends, preventing transpososome assembly. The model not only explains the in vivo transposase dose-response for Hsmar1 but also for the related Sleeping Beauty (SB) and piggyBac (PB) transposons. As yet, no information is at present available concerning the relevance of this mode of regulation to prokaryotic transposable elements.
Bibliography
- ↑ Machida C, Machida Y . Regulation of IS1 transposition by the insA gene product. - J Mol Biol: 1989 Aug 20, 208(4);567-74 [PubMed:2553980] [DOI]
- ↑ 2.0 2.1 Escoubas JM, Prère MF, Fayet O, Salvignol I, Galas D, Zerbib D, Chandler M . Translational control of transposition activity of the bacterial insertion sequence IS1. - EMBO J: 1991 Mar, 10(3);705-12 [PubMed:1848178] [DOI]
- ↑ Hu ST, Hwang JH, Lee LC, Lee CH, Li PL, Hsieh YC . Functional analysis of the 14 kDa protein of insertion sequence 2. - J Mol Biol: 1994 Feb 18, 236(2);503-13 [PubMed:8107136] [DOI]
- ↑ 4.0 4.1 4.2 Kleckner N, Chalmers RM, Kwon D, Sakai J, Bolland S . Tn10 and IS10 transposition and chromosome rearrangements: mechanism and regulation in vivo and in vitro. - Curr Top Microbiol Immunol: 1996, 204;49-82 [PubMed:8556869] [DOI]
- ↑ 5.0 5.1 Machida Y, Machida C, Ohtsubo H, Ohtsubo E . Factors determining frequency of plasmid cointegration mediated by insertion sequence IS1. - Proc Natl Acad Sci U S A: 1982 Jan, 79(2);277-81 [PubMed:6281761] [DOI]
- ↑ Chandler M, Galas DJ . Cointegrate formation mediated by Tn9. II. Activity of IS1 is modulated by external DNA sequences. - J Mol Biol: 1983 Oct 15, 170(1);61-91 [PubMed:6313938] [DOI]
- ↑ 7.0 7.1 Goosen N, van de Putte P . Role of ner protein in bacteriophage Mu transposition. - J Bacteriol: 1986 Aug, 167(2);503-7 [PubMed:3015876] [DOI]
- ↑ Davis MA, Simons RW, Kleckner N . Tn10 protects itself at two levels from fortuitous activation by external promoters. - Cell: 1985 Nov, 43(1);379-87 [PubMed:2416461] [DOI]
- ↑ 9.0 9.1 Krebs MP, Reznikoff WS . Transcriptional and translational initiation sites of IS50. Control of transposase and inhibitor expression. - J Mol Biol: 1986 Dec 20, 192(4);781-91 [PubMed:2438419] [DOI]
- ↑ Rezsöhazy R, Hallet B, Delcour J, Mahillon J . The IS4 family of insertion sequences: evidence for a conserved transposase motif. - Mol Microbiol: 1993 Sep, 9(6);1283-95 [PubMed:7934941] [DOI]
- ↑ 11.0 11.1 Chandler M, Fayet O . Translational frameshifting in the control of transposition in bacteria. - Mol Microbiol: 1993 Feb, 7(4);497-503 [PubMed:8384687] [DOI]
- ↑ Farabaugh PJ . Programmed translational frameshifting. - Microbiol Rev: 1996 Mar, 60(1);103-34 [PubMed:8852897] [DOI]
- ↑ Gesteland RF, Atkins JF . Recoding: dynamic reprogramming of translation. - Annu Rev Biochem: 1996, 65;741-68 [PubMed:8811194] [DOI]
- ↑ Sekine Y, Ohtsubo E . Frameshifting is required for production of the transposase encoded by insertion sequence 1. - Proc Natl Acad Sci U S A: 1989 Jun, 86(12);4609-13 [PubMed:2543983] [DOI]
- ↑ Polard P, Prère MF, Chandler M, Fayet O . Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. - J Mol Biol: 1991 Dec 5, 222(3);465-77 [PubMed:1660923] [DOI]
- ↑ Baranov PV, Hammer AW, Zhou J, Gesteland RF, Atkins JF . Transcriptional slippage in bacteria: distribution in sequenced genomes and utilization in IS element gene expression. - Genome Biol: 2005, 6(3);R25 [PubMed:15774026] [DOI]
- ↑ Baranov PV, Fayet O, Hendrix RW, Atkins JF . Recoding in bacteriophages and bacterial IS elements. - Trends Genet: 2006 Mar, 22(3);174-81 [PubMed:16460832] [DOI]
- ↑ Sharma V, Firth AE, Antonov I, Fayet O, Atkins JF, Borodovsky M, Baranov PV . A pilot study of bacterial genes with disrupted ORFs reveals a surprising profusion of protein sequence recoding mediated by ribosomal frameshifting and transcriptional realignment. - Mol Biol Evol: 2011 Nov, 28(11);3195-211 [PubMed:21673094] [DOI]
- ↑ Paul L, Ferguson DJ Jr, Krzycki JA . The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. - J Bacteriol: 2000 May, 182(9);2520-9 [PubMed:10762254] [DOI]
- ↑ Zhang Y, Baranov PV, Atkins JF, Gladyshev VN . Pyrrolysine and selenocysteine use dissimilar decoding strategies. - J Biol Chem: 2005 May 27, 280(21);20740-51 [PubMed:15788401] [DOI]
- ↑ Prat L, Heinemann IU, Aerni HR, Rinehart J, O'Donoghue P, Söll D . Carbon source-dependent expansion of the genetic code in bacteria. - Proc Natl Acad Sci U S A: 2012 Dec 18, 109(51);21070-5 [PubMed:23185002] [DOI]
- ↑ Schimmel P, Beebe K . Molecular biology: genetic code seizes pyrrolysine. - Nature: 2004 Sep 16, 431(7006);257-8 [PubMed:15372017] [DOI]
- ↑ Srinivasan G, James CM, Krzycki JA . Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. - Science: 2002 May 24, 296(5572);1459-62 [PubMed:12029131] [DOI]
- ↑ De Meirsman C, Van Soom C, Verreth C, Van Gool A, Vanderleyden J . Nucleotide sequence analysis of IS427 and its target sites in Agrobacterium tumefaciens T37. - Plasmid: 1990 Nov, 24(3);227-34 [PubMed:1963949] [DOI]
- ↑ Mariani F, Piccolella E, Colizzi V, Rappuoli R, Gross R . Characterization of an IS-like element from Mycobacterium tuberculosis. - J Gen Microbiol: 1993 Aug, 139(8);1767-72 [PubMed:8409920] [DOI]
- ↑ Fournier P, Paulus F, Otten L . IS870 requires a 5'-CTAG-3' target sequence to generate the stop codon for its large ORF1. - J Bacteriol: 1993 May, 175(10);3151-60 [PubMed:8387998] [DOI]
- ↑ 27.0 27.1 27.2 27.3 Derbyshire KM, Kramer M, Grindley ND . Role of instability in the cis action of the insertion sequence IS903 transposase. - Proc Natl Acad Sci U S A: 1990 Jun, 87(11);4048-52 [PubMed:2161528] [DOI]
- ↑ 28.0 28.1 Derbyshire KM, Grindley ND . Cis preference of the IS903 transposase is mediated by a combination of transposase instability and inefficient translation. - Mol Microbiol: 1996 Sep, 21(6);1261-72 [PubMed:8898394] [DOI]
- ↑ Reif HJ, Saedler H . IS1 is involved in deletion formation in the gal region of E. coli K12. - Mol Gen Genet: 1975, 137(1);17-28 [PubMed:1101028] [DOI]
- ↑ Kretschmer PJ, Cohen SN . Effect of temperature on translocation frequency of the Tn3 element. - J Bacteriol: 1979 Aug, 139(2);515-9 [PubMed:378977] [DOI]
- ↑ Haren L, Bétermier M, Polard P, Chandler M . IS911-mediated intramolecular transposition is naturally temperature sensitive. - Mol Microbiol: 1997 Aug, 25(3);531-40 [PubMed:9302015] [DOI]
- ↑ Gueguen E, Rousseau P, Duval-Valentin G, Chandler M . Truncated forms of IS911 transposase downregulate transposition. - Mol Microbiol: 2006 Nov, 62(4);1102-16 [PubMed:17078817] [DOI]
- ↑ Morisato D, Way JC, Kim HJ, Kleckner N . Tn10 transposase acts preferentially on nearby transposon ends in vivo. - Cell: 1983 Mar, 32(3);799-807 [PubMed:6299577] [DOI]
- ↑ Isberg RR, Lazaar AL, Syvanen M . Regulation of Tn5 by the right-repeat proteins: control at the level of the transposition reaction? - Cell: 1982 Oct, 30(3);883-92 [PubMed:6291787] [DOI]
- ↑ 35.0 35.1 Grindley ND, Joyce CM . Analysis of the structure and function of the kanamycin-resistance transposon Tn903. - Cold Spring Harb Symp Quant Biol: 1981, 45 Pt 1;125-33 [PubMed:6271455] [DOI]
- ↑ 36.0 36.1 Duval-Valentin G, Chandler M . Cotranslational control of DNA transposition: a window of opportunity. - Mol Cell: 2011 Dec 23, 44(6);989-96 [PubMed:22195971] [DOI]
- ↑ Nagy Z, Chandler M . Regulation of transposition in bacteria. - Res Microbiol: 2004 Jun, 155(5);387-98 [PubMed:15207871] [DOI]
- ↑ Jain C, Kleckner N . IS10 mRNA stability and steady state levels in Escherichia coli: indirect effects of translation and role of rne function. - Mol Microbiol: 1993 Jul, 9(2);233-47 [PubMed:7692216] [DOI]
- ↑ Jain C, Kleckner N . Preferential cis action of IS10 transposase depends upon its mode of synthesis. - Mol Microbiol: 1993 Jul, 9(2);249-60 [PubMed:8412678] [DOI]
- ↑ Sasakawa C, Lowe JB, McDivitt L, Berg DE . Control of transposon Tn5 transposition in Escherichia coli. - Proc Natl Acad Sci U S A: 1982 Dec, 79(23);7450-4 [PubMed:6296834] [DOI]
- ↑ Zerbib D, Jakowec M, Prentki P, Galas DJ, Chandler M . Expression of proteins essential for IS1 transposition: specific binding of InsA to the ends of IS1. - EMBO J: 1987 Oct, 6(10);3163-9 [PubMed:2826132] [DOI]
- ↑ Nagy Z, Szabó M, Chandler M, Olasz F . Analysis of the N-terminal DNA binding domain of the IS30 transposase. - Mol Microbiol: 2004 Oct, 54(2);478-88 [PubMed:15469518] [DOI]
- ↑ Stalder R, Caspers P, Olasz F, Arber W . The N-terminal domain of the insertion sequence 30 transposase interacts specifically with the terminal inverted repeats of the element. - J Biol Chem: 1990 Mar 5, 265(7);3757-62 [PubMed:2154486]
- ↑ Haren L, Normand C, Polard P, Alazard R, Chandler M . IS911 transposition is regulated by protein-protein interactions via a leucine zipper motif. - J Mol Biol: 2000 Feb 25, 296(3);757-68 [PubMed:10677279] [DOI]
- ↑ Normand C, Duval-Valentin G, Haren L, Chandler M . The terminal inverted repeats of IS911: requirements for synaptic complex assembly and activity. - J Mol Biol: 2001 May 18, 308(5);853-71 [PubMed:11352577] [DOI]
- ↑ Haren L, Polard P, Ton-Hoang B, Chandler M . Multiple oligomerisation domains in the IS911 transposase: a leucine zipper motif is essential for activity. - J Mol Biol: 1998, 283(1);29-41 [PubMed:9761671] [DOI]
- ↑ Chalmers RM, Kleckner N . Tn10/IS10 transposase purification, activation, and in vitro reaction. - J Biol Chem: 1994 Mar 18, 269(11);8029-35 [PubMed:8132525]
- ↑ Lewis LA, Astatke M, Umekubo PT, Alvi S, Saby R, Afrose J . Soluble expression, purification and characterization of the full length IS2 Transposase. - Mob DNA: 2011 Oct 27, 2;14 [PubMed:22032517] [DOI]
- ↑ Lewis LA, Astatke M, Umekubo PT, Alvi S, Saby R, Afrose J, Oliveira PH, Monteiro GA, Prazeres DM . Protein-DNA interactions define the mechanistic aspects of circle formation and insertion reactions in IS2 transposition. - Mob DNA: 2012 Jan 26, 3(1);1 [PubMed:22277150] [DOI]
- ↑ Lavoie BD, Chan BS, Allison RG, Chaconas G . Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. - EMBO J: 1991 Oct, 10(10);3051-9 [PubMed:1655409] [DOI]
- ↑ Mahnke Braam LA, Goryshin IY, Reznikoff WS . A mechanism for Tn5 inhibition. carboxyl-terminal dimerization. - J Biol Chem: 1999 Jan 1, 274(1);86-92 [PubMed:9867814] [DOI]
- ↑ Reznikoff WS . Transposon Tn5. - Annu Rev Genet: 2008, 42;269-86 [PubMed:18680433] [DOI]
- ↑ Bao KK, Wang H, Miller JK, Erie DA, Skalka AM, Wong I . Functional oligomeric state of avian sarcoma virus integrase. - J Biol Chem: 2003 Jan 10, 278(2);1323-7 [PubMed:12446721] [DOI]
- ↑ Ren G, Gao K, Bushman FD, Yeager M . Single-particle image reconstruction of a tetramer of HIV integrase bound to DNA. - J Mol Biol: 2007 Feb 9, 366(1);286-94 [PubMed:17157316] [DOI]
- ↑ Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P . Structural basis for functional tetramerization of lentiviral integrase. - PLoS Pathog: 2009 Jul, 5(7);e1000515 [PubMed:19609359] [DOI]
- ↑ Michel F, Crucifix C, Granger F, Eiler S, Mouscadet JF, Korolev S, Agapkina J, Ziganshin R, Gottikh M, Nazabal A, Emiliani S, Benarous R, Moras D, Schultz P, Ruff M . Structural basis for HIV-1 DNA integration in the human genome, role of the LEDGF/P75 cofactor. - EMBO J: 2009 Apr 8, 28(7);980-91 [PubMed:19229293] [DOI]
- ↑ Shieh YW, Minguez P, Bork P, Auburger JJ, Guilbride DL, Kramer G, Bukau B . Operon structure and cotranslational subunit association direct protein assembly in bacteria. - Science: 2015 Nov 6, 350(6261);678-80 [PubMed:26405228] [DOI]
- ↑ Montaño SP, Pigli YZ, Rice PA . The μ transpososome structure sheds light on DDE recombinase evolution. - Nature: 2012 Nov 15, 491(7424);413-7 [PubMed:23135398] [DOI]
- ↑ Davies DR, Goryshin IY, Reznikoff WS, Rayment I . Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. - Science: 2000 Jul 7, 289(5476);77-85 [PubMed:10884228] [DOI]
- ↑ Chaconas G, Lavoie BD, Watson MA . DNA transposition: jumping gene machine, some assembly required. - Curr Biol: 1996 Jul 1, 6(7);817-20 [PubMed:8805293] [DOI]
- ↑ Gamas P, Galas D, Chandler M . DNA sequence at the end of IS1 required for transposition. - Nature: 1985 Oct 3-9, 317(6036);458-60 [PubMed:2995832] [DOI]
- ↑ Grindley ND, Joyce CM . Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. - Proc Natl Acad Sci U S A: 1980 Dec, 77(12);7176-80 [PubMed:6261245] [DOI]
- ↑ Signon L, Kleckner N . Negative and positive regulation of Tn10/IS10-promoted recombination by IHF: two distinguishable processes inhibit transposition off of multicopy plasmid replicons and activate chromosomal events that favor evolution of new transposons. - Genes Dev: 1995 May 1, 9(9);1123-36 [PubMed:7744253] [DOI]
- ↑ 64.0 64.1 Chalmers R, Guhathakurta A, Benjamin H, Kleckner N . IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. - Cell: 1998 May 29, 93(5);897-908 [PubMed:9630232] [DOI]
- ↑ Sakai J, Chalmers RM, Kleckner N . Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. - EMBO J: 1995 Sep 1, 14(17);4374-83 [PubMed:7556079] [DOI]
- ↑ Wiater LA, Grindley ND . Gamma delta transposase and integration host factor bind cooperatively at both ends of gamma delta. - EMBO J: 1988 Jun, 7(6);1907-11 [PubMed:2844529] [DOI]
- ↑ Crellin P, Chalmers R . Protein-DNA contacts and conformational changes in the Tn10 transpososome during assembly and activation for cleavage. - EMBO J: 2001 Jul 16, 20(14);3882-91 [PubMed:11447129] [DOI]
- ↑ Sewitz S, Crellin P, Chalmers R . The positive and negative regulation of Tn10 transposition by IHF is mediated by structurally asymmetric transposon arms. - Nucleic Acids Res: 2003 Oct 15, 31(20);5868-76 [PubMed:14530435] [DOI]
- ↑ Crellin P, Sewitz S, Chalmers R . DNA looping and catalysis; the IHF-folded arm of Tn10 promotes conformational changes and hairpin resolution. - Mol Cell: 2004 Feb 27, 13(4);537-47 [PubMed:14992723] [DOI]
- ↑ 70.0 70.1 Wardle SJ, Chan A, Haniford DB . H-NS binds with high affinity to the Tn10 transpososome and promotes transpososome stabilization. - Nucleic Acids Res: 2009 Oct, 37(18);6148-60 [PubMed:19696075] [DOI]
- ↑ Whitfield CR, Wardle SJ, Haniford DB . The global bacterial regulator H-NS promotes transpososome formation and transposition in the Tn5 system. - Nucleic Acids Res: 2009 Feb, 37(2);309-21 [PubMed:19042975] [DOI]
- ↑ 72.0 72.1 72.2 72.3 Wardle SJ, O'Carroll M, Derbyshire KM, Haniford DB . The global regulator H-NS acts directly on the transpososome to promote Tn10 transposition. - Genes Dev: 2005 Sep 15, 19(18);2224-35 [PubMed:16166383] [DOI]
- ↑ 73.0 73.1 Ward CM, Wardle SJ, Singh RK, Haniford DB . The global regulator H-NS binds to two distinct classes of sites within the Tn10 transpososome to promote transposition. - Mol Microbiol: 2007 May, 64(4);1000-13 [PubMed:17501923] [DOI]
- ↑ Reznikoff WS . The Tn5 transposon. - Annu Rev Microbiol: 1993, 47;945-63 [PubMed:7504907] [DOI]
- ↑ Shiga Y, Sekine Y, Ohtsubo E . Transposition of IS1 circles. - Genes Cells: 1999 Oct, 4(10);551-61 [PubMed:10583504] [DOI]
- ↑ 76.0 76.1 Maekawa T, Yanagihara K, Ohtsubo E . Specific nicking at the 3' ends of the terminal inverted repeat sequences in transposon Tn3 by transposase and an E. coli protein ACP. - Genes Cells: 1996 Nov, 1(11);1017-30 [PubMed:9077464] [DOI]
- ↑ 77.0 77.1 Hallet B, Sherratt DJ . Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. - FEMS Microbiol Rev: 1997 Sep, 21(2);157-78 [PubMed:9348666] [DOI]
- ↑ 78.0 78.1 Sharpe PL, Craig NL . Host proteins can stimulate Tn7 transposition: a novel role for the ribosomal protein L29 and the acyl carrier protein. - EMBO J: 1998 Oct 1, 17(19);5822-31 [PubMed:9755182] [DOI]
- ↑ Mhammedi-Alaoui A, Pato M, Gama MJ, Toussaint A . A new component of bacteriophage Mu replicative transposition machinery: the Escherichia coli ClpX protein. - Mol Microbiol: 1994 Mar, 11(6);1109-16 [PubMed:8022280] [DOI]
- ↑ Kruklitis R, Welty DJ, Nakai H . ClpX protein of Escherichia coli activates bacteriophage Mu transposase in the strand transfer complex for initiation of Mu DNA synthesis. - EMBO J: 1996 Feb 15, 15(4);935-44 [PubMed:8631314]
- ↑ Levchenko I, Luo L, Baker TA . Disassembly of the Mu transposase tetramer by the ClpX chaperone. - Genes Dev: 1995 Oct 1, 9(19);2399-408 [PubMed:7557391] [DOI]
- ↑ Levchenko I, Yamauchi M, Baker TA . ClpX and MuB interact with overlapping regions of Mu transposase: implications for control of the transposition pathway. - Genes Dev: 1997 Jun 15, 11(12);1561-72 [PubMed:9203582] [DOI]
- ↑ Laachouch JE, Desmet L, Geuskens V, Grimaud R, Toussaint A . Bacteriophage Mu repressor as a target for the Escherichia coli ATP-dependent Clp Protease. - EMBO J: 1996 Jan 15, 15(2);437-44 [PubMed:8617219]
- ↑ Welty DJ, Jones JM, Nakai H . Communication of ClpXP protease hypersensitivity to bacteriophage Mu repressor isoforms. - J Mol Biol: 1997 Sep 12, 272(1);31-41 [PubMed:9299335] [DOI]
- ↑ Lane D, Cavaillé J, Chandler M . Induction of the SOS response by IS1 transposase. - J Mol Biol: 1994 Sep 30, 242(4);339-50 [PubMed:7932694] [DOI]
- ↑ Mahillon J, Chandler M . Insertion sequences. - Microbiol Mol Biol Rev: 1998 Sep, 62(3);725-74 [PubMed:9729608] [DOI]
- ↑ 87.0 87.1 Kuan CT, Liu SK, Tessman I . Excision and transposition of Tn5 as an SOS activity in Escherichia coli. - Genetics: 1991 May, 128(1);45-57 [PubMed:1648004] [DOI]
- ↑ 88.0 88.1 Kuan CT, Tessman I . LexA protein of Escherichia coli represses expression of the Tn5 transposase gene. - J Bacteriol: 1991 Oct, 173(20);6406-10 [PubMed:1655708] [DOI]
- ↑ 89.0 89.1 Weinreich MD, Makris JC, Reznikoff WS . Induction of the SOS response in Escherichia coli inhibits Tn5 and IS50 transposition. - J Bacteriol: 1991 Nov, 173(21);6910-8 [PubMed:1657870] [DOI]
- ↑ Kuan CT, Tessman I . Further evidence that transposition of Tn5 in Escherichia coli is strongly enhanced by constitutively activated RecA proteins. - J Bacteriol: 1992 Nov, 174(21);6872-7 [PubMed:1328165] [DOI]
- ↑ 91.0 91.1 91.2 Ahmed A . Evidence for replicative transposition of Tn5 and Tn9. - J Mol Biol: 1986 Sep 5, 191(1);75-84 [PubMed:3025455] [DOI]
- ↑ Nagel R, Chan A . Enhanced Tn10 and mini-Tn10 precise excision in DNA replication mutants of Escherichia coli K12. - Mutat Res: 2000 May 31, 459(4);275-84 [PubMed:10844241] [DOI]
- ↑ Reddy M, Gowrishankar J . Characterization of the uup locus and its role in transposon excisions and tandem repeat deletions in Escherichia coli. - J Bacteriol: 2000 Apr, 182(7);1978-86 [PubMed:10715006] [DOI]
- ↑ Sasakawa C, Uno Y, Yoshikawa M . The requirement for both DNA polymerase and 5' to 3' exonuclease activities of DNA polymerase I during Tn5 transposition. - Mol Gen Genet: 1981, 182(1);19-24 [PubMed:6267432] [DOI]
- ↑ Syvanen M, Hopkins JD, Clements M . A new class of mutants in DNA polymerase I that affects gene transposition. - J Mol Biol: 1982 Jun 25, 158(2);203-12 [PubMed:6288966] [DOI]
- ↑ Sasakawa C, Uno Y, Yoshikawa M . lon-sulA regulatory function affects the efficiency of transposition of Tn5 from lambda b221 cI857 Pam Oam to the chromosome. - Biochem Biophys Res Commun: 1987 Feb 13, 142(3);879-84 [PubMed:3030303] [DOI]
- ↑ Isberg RR, Syvanen M . DNA gyrase is a host factor required for transposition of Tn5. - Cell: 1982 Aug, 30(1);9-18 [PubMed:6290084] [DOI]
- ↑ Sternglanz R, DiNardo S, Voelkel KA, Nishimura Y, Hirota Y, Becherer K, Zumstein L, Wang JC . Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. - Proc Natl Acad Sci U S A: 1981 May, 78(5);2747-51 [PubMed:6265907] [DOI]
- ↑ Pato ML, Karlok M, Wall C, Higgins NP . Characterization of Mu prophage lacking the central strong gyrase binding site: localization of the block in replication. - J Bacteriol: 1995 Oct, 177(20);5937-42 [PubMed:7592347] [DOI]
- ↑ Pato ML, Banerjee M . The Mu strong gyrase-binding site promotes efficient synapsis of the prophage termini. - Mol Microbiol: 1996 Oct, 22(2);283-92 [PubMed:8930913] [DOI]
- ↑ 101.0 101.1 Roberts D, Hoopes BC, McClure WR, Kleckner N . IS10 transposition is regulated by DNA adenine methylation. - Cell: 1985 Nov, 43(1);117-30 [PubMed:3000598] [DOI]
- ↑ Yin JC, Krebs MP, Reznikoff WS . Effect of dam methylation on Tn5 transposition. - J Mol Biol: 1988 Jan 5, 199(1);35-45 [PubMed:2451025] [DOI]
- ↑ Davies DR, Mahnke Braam L, Reznikoff WS, Rayment I . The three-dimensional structure of a Tn5 transposase-related protein determined to 2.9-A resolution. - J Biol Chem: 1999 Apr 23, 274(17);11904-13 [PubMed:10207011] [DOI]
- ↑ Twiss E, Coros AM, Tavakoli NP, Derbyshire KM . Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress. - Mol Microbiol: 2005 Sep, 57(6);1593-607 [PubMed:16135227] [DOI]
- ↑ Coros AM, Twiss E, Tavakoli NP, Derbyshire KM . Genetic evidence that GTP is required for transposition of IS903 and Tn552 in Escherichia coli. - J Bacteriol: 2005 Jul, 187(13);4598-606 [PubMed:15968071] [DOI]
- ↑ Ross JA, Ellis MJ, Hossain S, Haniford DB . Hfq restructures RNA-IN and RNA-OUT and facilitates antisense pairing in the Tn10/IS10 system. - RNA: 2013 May, 19(5);670-84 [PubMed:23510801] [DOI]
- ↑ Ellis MJ, Trussler RS, Haniford DB . Hfq binds directly to the ribosome-binding site of IS10 transposase mRNA to inhibit translation. - Mol Microbiol: 2015 May, 96(3);633-50 [PubMed:25649688] [DOI]
- ↑ Ellis MJ, Trussler RS, Ross JA, Haniford DB . Probing Hfq:RNA interactions with hydroxyl radical and RNase footprinting. - Methods Mol Biol: 2015, 1259;403-15 [PubMed:25579599] [DOI]
- ↑ Lorenzetti APR, Kusebauch U, Zaramela LS, Wu WJ, de Almeida JPP, Turkarslan S, L G de Lomana A, Gomes-Filho JV, Vêncio RZN, Moritz RL, Koide T, Baliga NS . A Genome-Scale Atlas Reveals Complex Interplay of Transcription and Translation in an Archaeon. - mSystems: 2023 Apr 27, 8(2);e0081622 [PubMed:36912639] [DOI]
- ↑ Lampe DJ, Grant TE, Robertson HM . Factors affecting transposition of the Himar1 mariner transposon in vitro. - Genetics: 1998 May, 149(1);179-87 [PubMed:9584095] [DOI]
- ↑ Lohe AR, Hartl DL . Autoregulation of mariner transposase activity by overproduction and dominant-negative complementation. - Mol Biol Evol: 1996 Apr, 13(4);549-55 [PubMed:8882498] [DOI]
- ↑ Hartl DL, Lozovskaya ER, Nurminsky DI, Lohe AR . What restricts the activity of mariner-like transposable elements. - Trends Genet: 1997 May, 13(5);197-201 [PubMed:9154003] [DOI]
- ↑ Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, Bell JB, Largaespada DA, Hackett PB . Gene transfer into genomes of human cells by the sleeping beauty transposon system. - Mol Ther: 2003 Jul, 8(1);108-17 [PubMed:12842434] [DOI]
- ↑ Zayed H, Izsvák Z, Walisko O, Ivics Z . Development of hyperactive sleeping beauty transposon vectors by mutational analysis. - Mol Ther: 2004 Feb, 9(2);292-304 [PubMed:14759813] [DOI]
- ↑ Karsi A, Moav B, Hackett P, Liu Z . Effects of insert size on transposition efficiency of the sleeping beauty transposon in mouse cells. - Mar Biotechnol (NY): 2001 May, 3(3);241-5 [PubMed:14961361] [DOI]
- ↑ Mikkelsen JG, Yant SR, Meuse L, Huang Z, Xu H, Kay MA . Helper-Independent Sleeping Beauty transposon-transposase vectors for efficient nonviral gene delivery and persistent gene expression in vivo. - Mol Ther: 2003 Oct, 8(4);654-65 [PubMed:14529839] [DOI]
- ↑ Claeys Bouuaert C, Lipkow K, Andrews SS, Liu D, Chalmers R . The autoregulation of a eukaryotic DNA transposon. - Elife: 2013 Jun 18, 2;e00668 [PubMed:23795293] [DOI]
- ↑ Bouuaert CC, Tellier M, Chalmers R . One to rule them all: A highly conserved motif in mariner transposase controls multiple steps of transposition. - Mob Genet Elements: 2014 Jan 1, 4(1);e28807 [PubMed:24812590] [DOI]
- ↑ Tellier M, Bouuaert CC, Chalmers R . Mariner and the ITm Superfamily of Transposons. - Microbiol Spectr: 2015 Apr, 3(2);MDNA3-0033-2014. [PubMed:26104691] [DOI]