IS Families/IS3 family
Contents
- 1 Original Identification
- 2 Presence in Compound Transposons
- 3 Distribution
- 4 Organization
- 5 Formation of a strong transposase promoter
- 6 Regulation by Methylation?
- 7 Insertion specificity
- 8 Group II intron insertions
- 9 IS3 family subgroups
- 10 Family Exceptions
- 11 Mycoplasma and the non-universal genetic code
- 12 A clade with non-canonical IR
- 13 An additional subgroup
- 14 Mechanism
- 14.1 Transposition Proteins
- 14.2 OrfA
- 14.3 OrfB
- 14.4 OrfAB: a product of programmed ribosomal frameshifting (PRTF)
- 14.5 Artificial orfA-orfB fusion
- 14.6 Structural motifs
- 14.7 Co-translational DNA binding
- 14.8 Co-translational multimerisation
- 14.9 The IS911 transpososome
- 14.10 Excision synaptic complex SCA.
- 14.11 Insertion synaptic complex SCB
- 14.12 The Transposition Pathway
- 14.13 The Figure-eight form
- 14.14 The circular intermediate
- 14.15 Integration of the circular intermediate
- 14.16 Targeted Insertion
- 14.17 Mechanism in other family members
- 15 Structural studies
- 16 Excision: A dedicated enzyme
- 16.1 IS Excision is Stimulated by High Transposase Levels
- 16.2 A Dedicated Enzyme: Identification of a common reading frame ECs1305 in all high excision strains
- 16.3 ECs1305 and an active transposase is required for high level IS excision
- 16.4 Analysis of the primary Sequence of IEE: a PrimPol Helicase
- 16.5 Biochemical Analysis of IEE Activities: Polymerase Activity
- 16.6 Microhomology-Mediated End-Joining
- 16.7 The C-terminal Helicase Domain
- 16.8 Models for IEE activity
- 16.9 The mcr connection: a model for excision by replication-associated strand exchange and primer invasion.
- 16.10 The Primer Invasion model also explains high levels of precise excision.
- 16.11 IEE and the Diagnostics Lab
- 17 Acknowledgements
- 18 Bibliography
Original Identification
IS3 and another member of this family, IS2 were identified genetically as a DNA segments causing insertional inactivation of gal and lac operons and physically by electron microscopy[1] and in plasmid F as a segment called alpha-beta[2][3]. IS3 was subsequently wrongly identified as the insertion sequence flanking the tetracycline resistance transposon Tn10[4][5]. It has subsequently been found as a component of a large number of plasmids particularly in gram negative enterics.
Presence in Compound Transposons
Although IS3 family elements do participate in compound transposons (e.g. IS3411) flanking the Citrate Utilization, to our knowledge there has been no systematic survey undertaken and very few IS3-associated compounds have been described to date. Several family members are part of compound transposons. These include: IS3411 flanking genes for citrate utilization in transposon Tn3411[6][7][8], IS4521 which flanks a heat stable enterotoxin gene in enterotoxinogenic Escherichia coli and IS1706, which flanks genes of the Clp protease/chaperone family.
Distribution
This is one of the most coherent, largest, most abundant and widely distributed IS families [9] (see [10]). Nearly 600 individual different members of this family have been identified in more than 267 bacterial species distributed over 145 genera. However, their true distribution is clearly significantly greater than this.
For example, IS911, (isolated from a Shigella dysenteriae phage λ lysogen by spontaneous insertion into the phage cI repressor gene[11]) is present in multiple copies in the original host strain and in type strains of other Shigella species. Two vestigial copies, both interrupted by a copy of IS30, were also detected in the chromosome of E. coli K12[12] and could form transposition intermediates when supplied with IS911 transposase[13]. Entire or truncated IS911 copies have also been identified in several E. coli virulence plasmids (e.g. [14]), in pathogenicity islands of uropathogenic E. coli (e.g. [15]), in various other clinical isolates of E. coli and in a large number of well-known and less well-known enterobacteria such as Escherichia fergusonii, Chronobacter, Dickeya, Erwinia, Klebsiella, Pantoea, Shimwellia, and Yersinia.
Most IS3 family members have been identified in bacteria although at least one example, ISMco1, has also been identified in the archaea Methanosaeta concilii[16]. Since this archaeon is widespread in nature[17], it is possible that this represents a case of recent horizontal transfer. The presence of 8 copies implies that ISMco1 is active in its archaeal host.
Organization
The family is quite homogenous in the organization (Fig.IS3.1). in spite of its wide distribution in bacteria exhibiting a large range of G+C contents (from 70% in the Mycobacterial examples to 25% in those isolated from Mycoplasma) and of the presence of members in hosts such as Mycoplasma with a non-universal genetic code (e.g. IS1138) or in bacteria which use stop codon read-through by insertion of the unusual amino acid selenocysteine (e.g. ISDvu3 from Desulfovibrio vulgaris). In the case of both copies of IS1138, which participates in high frequency rearrangements of the Mycoplasma pulmonis chromosome, the Tpase orf carries 11 UGA codons which are decoded as tryptophan[18].
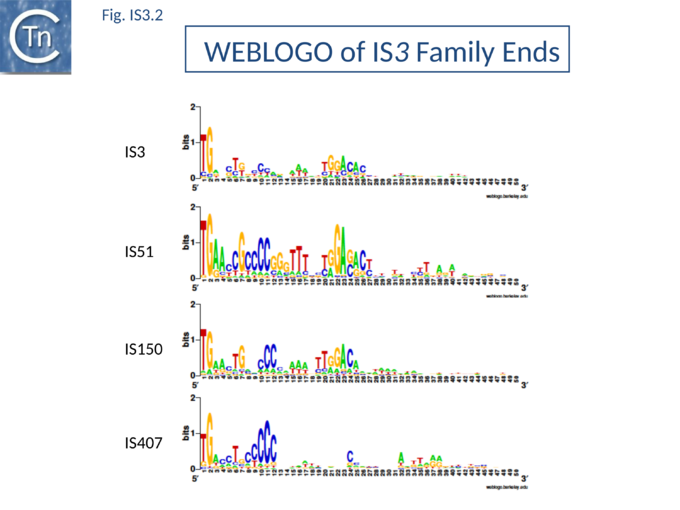
Members are between 1200 and 1550 bp with relatively well conserved inverted terminal repeats in the range of 20-40 bp. One exception previously attributed to this family, IS481, is 1045 bp long and has now been placed in a separate family; see "IS481 family"). They generate 3 or 4 bp DR on insertion.
The majority of IR terminate with 5'-TG-----CA-3' and present an internal block of G/C residues of variable length (Fig.IS3.2).
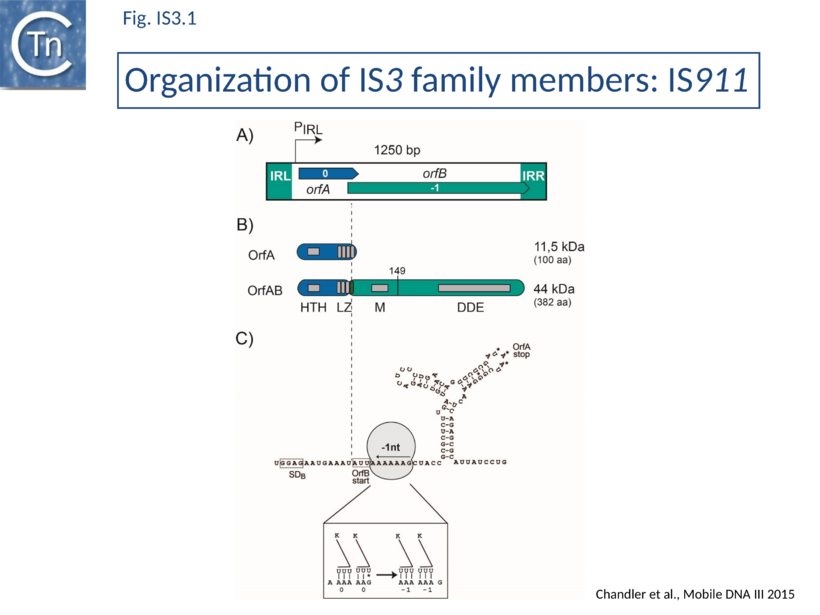
IS3-family members generally have two consecutive and partially overlapping reading frames, orfA and orfB, in relative translational reading phases 0 and -1, respectively (Fig.IS3.1 A) under control of a weak promoter, pIRL, partially located in IRL (Fig.IS3.1 A and Fig.IS3.3 C). The 5' end of orfB overlaps the 3' end of orfA and occurs in reading phase -1 relative to orfA (Fig.IS3.1).
It had been demonstrated in the 1990s that several family members (IS150[19], IS3[20], IS911[21], and IS2[22]) express two major proteins (Fig.IS3.1 B): OrfA, the product of the upstream frame,and the transposase, OrfAB, a “fusion” or “transframe” protein generated from orfA and orfB by Programmed -1 Ribosomal Frameshifting (PRF) (see "Programmed translational frameshifting")[23]. Many other members of this family are also organized in this way[24][25]. The frameshifting frequency varies from element to element. It is approximately 50% in the case of IS150[19] and only 15% for IS911[21].
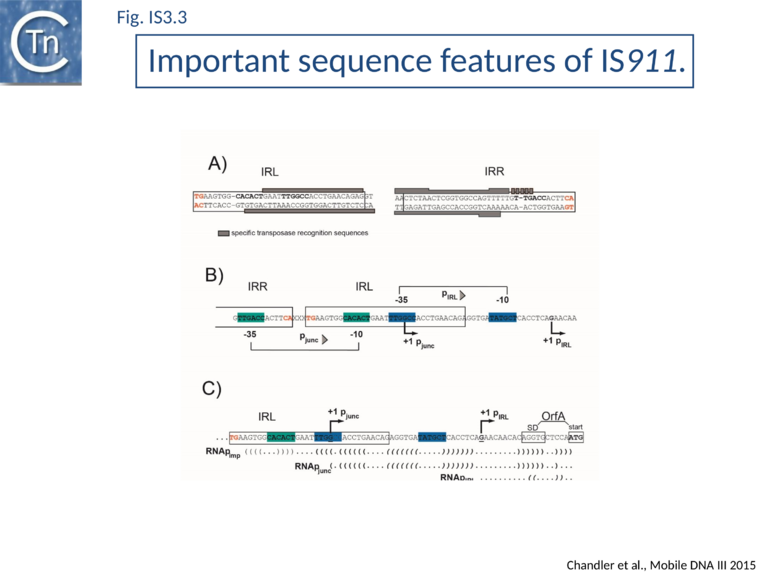
Complex internal inverted repeat sequences (Fig.IS3.3 C) (for IS911, located between coordinates 19 and 73) include the -35 and -10 hexamers of pIRL, the transcription start site and the ribosome binding site for OrfA. This is thought to play a role at the mRNA level in preventing excess transposase expression resulting from external transcription. The full secondary structure would be present in transcripts initiated outside the IS thus sequestering the translation initiation signals but only the 3’ part would be present if transcription initiates at pIRL. In this case, the translation initiation signals would be exposed. Initial studies (Prère and Fayet pers communication) have shown that translation from the longer transcript is very low but that deletion of its 5’ end to “liberate” the ribosome binding site (Fig.IS3.3 C) indeed results in a significant increase in translation. In the related IS2 element, a similar sequence appears to function as a DNA binding site for the OrfA protein which represses promoter activity but further studies are necessary to confirm this[26].
Formation of a strong transposase promoter
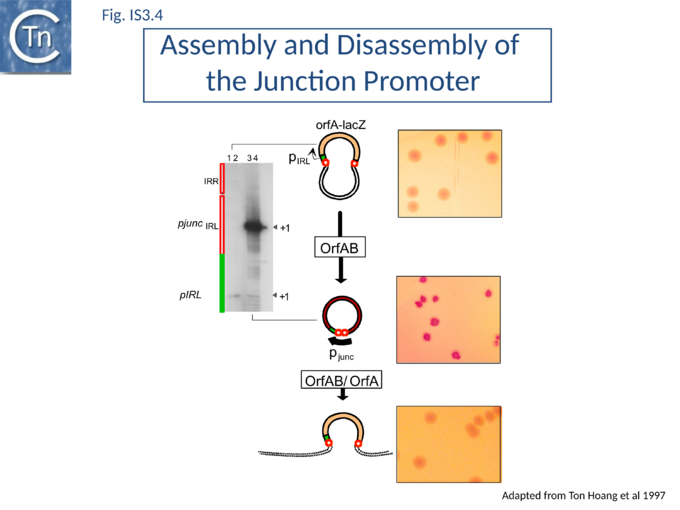
In common with many IS of other families (e.g. IS21[27], IS30[28], IS110[29][30]) the IS3 family IRR carry an outward-directed -35 promoter hexamer while IRL carries an inward-directed -10 promoter component (Fig.IS3.3 B). These are assembled into a strong promoter, pJunc, which serves to express high levels of transposition proteins (Fig.IS3.3 B); (Fig.IS3.4) in one of its key transposition intermediates, an excised transposon circle (see "Transposition Pathway"). Transcription initiation from pJunc, like that from impinging transcription, would also produce an RNA which could sequester the translation initiation signals but in a shorter and less stable stem loop structure (Fig.IS3.3 C).
Regulation by Methylation?
Several members carry GATC methylation sites within 50bp of their ends, which have been shown in one case, IS3, to modulate transposition activity[31], however, this is not a general characteristic of the family nor is it restricted to any particular subgroup.
Insertion specificity
There appears to be little sequence specificity for insertion of members of the family. IS2 exhibits a preference for a region of bacteriophage P1 but the basis of this preference is at present unknown[32]. Both IS911[33] and IS150 [34] have been found next to sequences which resemble their IRs (see “Targeted Insertion”) and IS1397 is invariably located within intergenic repeated sequences in E. coli (Bacterial Interspersed Mosaic Elements or BIMEs[35].
Group II intron insertions
Finally, an element isolated from the ECOR collection of E. coli and closely related to IS3411 carries a group II intron[36]. The effect of this on regulation of transposition of this element has not been investigated.
IS3 family subgroups
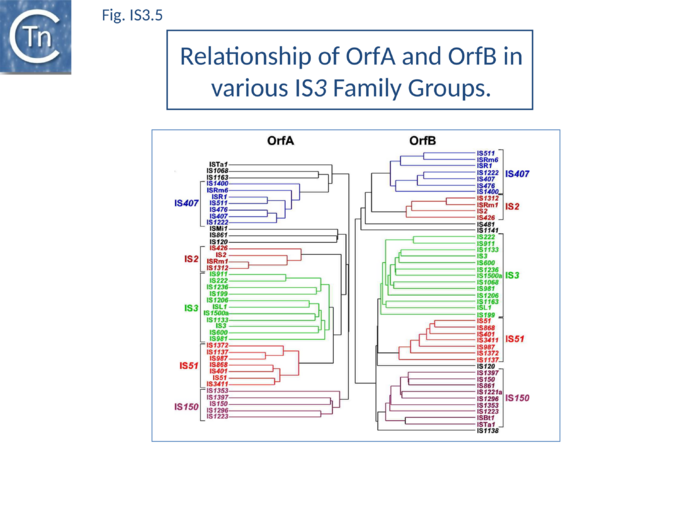
The IS3 family is divided into five subgroups (Table Characteristics of IS families; Fig.4.2). This is supported by deep branching in the alignment of the various OrfA and OrfB sequences[37] (Fig.IS3.5). These are: the IS2 and IS407 subgroups (which appear closely related), and the IS3, IS51, and IS150 subgroups.
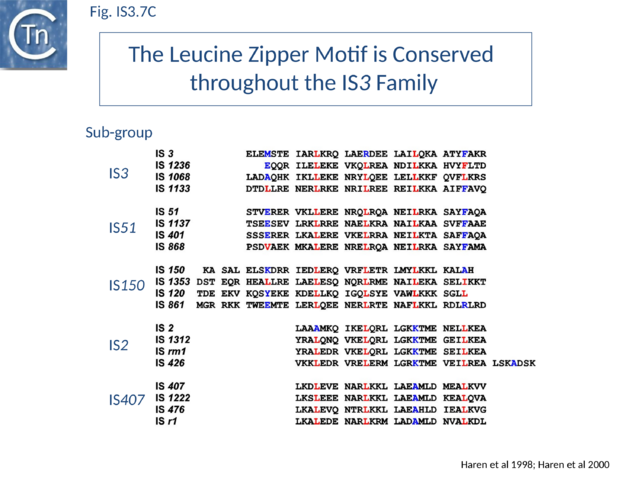
Additional members of the family identified subsequently also tend to follow this pattern. One feature which lends biological credence to these subgroups is that they also clearly appear clustered (with some exceptions) in the results of the alignments with the upstream OrfA protein[37]. Moreover, there is some correlation between the members of each group and the number of base pairs of target DNA duplicated on insertion (DR): for those elements in the IS2 subgroup, insertion invariably leads to a 5 bp DR; for the IS407 subgroup a 4 bp DR is observed; while for the other groups a 3 bp DR is generated (Table Characteristics of IS families). In the latter cases some of the elements, e.g. IS911, have been shown to occasionally generate 4 bp repeats. This clustering is also exhibited to some extent in the nucleotide sequence of the terminal IRs (Fig.IS3.2) and is particularly marked in the IS2, IS51 and IS407 subgroups. It can also be observed in the primary sequence details of the putative leucine zipper[38].
Family Exceptions
Several family members exhibit an organization which does not apparently conform to the generic IS3 member. In IS120, for example, the relationship between the reading phases of the upstream and downstream orfs appears to be +1 rather than -1 while in ISNg1 and ISYe1 the characteristic motifs of OrfB are distributed between reading phases. Other members, such as IS1076, IS1138, IS1221, and IS1141, exhibit only one long open reading frame. Although these may be true variants, it cannot at present be ruled out that the variations are simply due to errors in sequence determination.
Mycoplasma and the non-universal genetic code
Family members from Mycoplasma merit special attention. Not only does the host use a non-universal genetic code in which the opal termination codon TGA directs the insertion of tryptophan (see [39], but their genomes are among the smallest bacterial genomes known and extremely rich in A+T. To date, several different IS3 family members have been observed in Mycoplasma. Of these, only IS1138 (and IS1138b) has been demonstrated directly to undergo autonomous transposition[18]. All exhibit similarly high AT levels and this unusual base composition could lead to difficulties in sequence determination. It is remarkable that typical IS3 family characters have been maintained in such an "extreme" genetic environment. Nine individuals are closely related and form a group of iso-elements which have been called IS1221. As indicated above, one of these carries a single long reading frame (representing orfA + orfB) instead of two consecutive overlapping frames. The others each carry insertions or deletions which destroy either the equivalent of orfA, orfB, or both. Expression studies in E. coli indicate that a protein, equivalent to OrfAB, is indeed produced from the long open reading frame of IS1221. Interestingly, it appears that a second truncated protein, equivalent to OrfA, may be generated from the single orfAB frame by translational frameshifting, representing an "inverted" expression pattern to the majority of the family members[40]. Although this appears not to be a general rule for IS3 family members originating from Mycoplasma hosts, the presence of a similar single-frame arrangement in a second member, IS1138, indicates that it might not be rare. Because of the extremely high AT content of these elements, many potential frameshift windows of the A6G(/C) or A7 type are expected to occur. The only direct experiment will, therefore, be able to determine which, if any, of these sequences are used to generate the Tpase or, conversely, an OrfA-like protein.
A clade with non-canonical IR
A clade carrying non-canonical ends has recently been identified. These IS include 7 supplementary base pairs on each end flanking canonical IS3 ends: a conserved stretch of 5 C residues is located 5’ to the left IR and a less conserved motif (CGG) is located 3’ to the right end. When these additional bases are taken into account every member of this clade exhibits a 4 bp DR characteristic of the IS3 family (Table Characteristics of IS families) (Gourbeyre, pers. comm.). This conclusion is supported by the presence of multiple IS copies (e.g. ISPsy31) and also by identification of “empty sites”. This clearly requires further experimental investigation.
An additional subgroup
Recently, an additional subgroup has been proposed which includes ISPpy1[41]. However, all members belong to the IS150 subgroup and their Tpases are not separated by our standard multiple alignments and MCL analysis. Although they do exhibit some variation in the sequence of their terminal dinucleotides, similar variations are found for IS2 and members of other IS3 subgroups.
Mechanism
Transposition Proteins
Extensive alignment studies of the predicted OrfA and OrfB amino acid sequences between themselves and with those of other transposable elements[42][43][44][45][46] provided insights into structure/function relationships of the proteins (Fig.IS3.1 B).
OrfA
OrfA is small. For IS911 it has a predicted molecular weight of 11.5 kDa. The predicted primary amino acid sequences of most IS3 family members exhibit a similarly placed HTH signature (see for example [11][47]) which initially suggested that they might provide sequence-specific binding to the terminal IRs of their particular IS[48] involved in sequence-specific binding of the transposase to the terminal IRs OrfAB which was subsequently confirmed experimentally[49]. They also carry a C-terminal leucine zipper (LZ) motif first identified in IS2, IS150 and IS3 and which appears to be conserved in the majority of known members[50] and is involved in protein multimerization[11][21][38][50].
OrfB
The OrfB products carry a DD(35)E catalytic motif and share additional identities with retroviral integrases and various other Tpases[21][42][43][44][45][46][51]. These include two amino acids located 4 and 7 residues downstream from the glutamate residue.
IS911 OrfB is 299 residues long with a predicted molecular weight of 34.6kD. Its TAA termination codon lies just within IRR and may be significant in regulation. The OrfB initiation codon is AUU and consequently initiation occurs only at low levels[21][52] and is modulated by the level of initiation factor IF3[53].
OrfB has been observed for: IS3[20] (Prère & Fayet, unpublished), IS150[19], IS911[21][52][53] and IS3411/IS629[54][55] but not for IS2[56]. It is generally present at quite low levels although for IS3 approximately equal amounts of OrfB and OrfAB appear to be produced[20]. The IS150 OrfB initiation codon is out of phase with the rest of the gene and expression of full length OrfB would require a -1 frameshift after initiation.
Sequence analysis suggests that OrfB may in fact be synthesized by about 34% of IS3 family members through translational coupling: the stop codon of orfA overlaps with a potential orfB start codon (e.g. AUGA or GUGA) in 134 out of 399 ISs analyzed[24].
It is possible that the OrfB protein itself plays no direct role in transposition chemistry but that it is simply its translation signals which are important. Their recognition by the ribosome could modulate programmed translational frameshifting required to generate a single transposase protein, OrfAB, from the two reading frames orfA and orfB (see "Programmed translational frameshifting").
The OrfB amino acid sequence shares significant similarities with retroviral integrases, an observation which contributed to defining the highly conserved amino acid triad DDE common to all IS3 family members and to many of this type of phophoryltransferase enzymes[43][57]. This constitutes part of the active site (for reviews see: [48][52]).
OrfB carries neither the HTH nor the LZ motif.
OrfAB: a product of programmed ribosomal frameshifting (PRTF)
OrfAB is assembled from orfA and orfB by a programmed –1 ribosomal frameshift occurring near the 3' end of orfA (see "Programmed translational frameshifting") first demonstrated for the related IS150[19].
The transframe protein combines the orfA HTH motif, an LZ motif and the orfB DD(35)E catalytic domain [50] (Fig.IS3.1 B).
OrfAB of IS911 (382 amino acids) shares its 86 N-terminal amino acids with OrfA (100 amino acids) and its 296 C-terminal amino acids with OrfB (299 amino acids).
Ribosome rephasing to generate OrfAB occurs on a group of "slippery” lysine codons with a frequency of about 15% (measured using systems driven by two different promoters; T7p10 and ptac). OrfA is therefore normally expressed at significantly higher levels than OrfAB. Frameshifting permits the combination of different functional protein domains (Fig.IS3.1 C)..
IS3-family frameshifting is similar to that used in some retroviruses to generate the pol-gag "polyprotein"[58] and in the dnaX gene of E. coli to synthesize γ the sub-unit of DNA polymerase III[59].
The relevant IS911 sequences involved in frameshifting are shown in (Fig.IS3.1 C). Examples of frameshifting sequences from other members of the family are shown in Fig.IS3.6. The group of slippery lysine codons is A AAA AAG and is directly preceded by the AUU OrfB initiation codon. Since E. coli does not encode a tRNALys with a 3’UUC5’ anti-codon for AAG, both lysine codons are decoded by the same tRNALys with a 3’UUU5’ anticodon. Its pairing is weaker with a G at the wobble position[60] probably because modifications of U34 increase the rigidity of the anticodon[61]. The presence of an upstream RBS (GGAG sequence) and a downstream secondary structure (Y-shaped stem-loop) stimulates ribosome rephasing in the -1 direction. What drives frameshifting is probably the thermodynamically favorable re-pairing of the two tRNALys from codons AAA-AAG to codons AAA-AAA[59][62]. The stimulators likely have a mechanical effect bringing back in the register the ribosome and the mRNA after tRNA slippage. Different groups of codons have been observed to allow rephasing of the ribosome[25] and, although the most common motif is A6G, different members of the IS3 family carry a variety of these (e.g. A3G for IS3; see Atkins & Gesteland, Recoding: expansion of decoding rules enriches gene expression, Springer 2010).
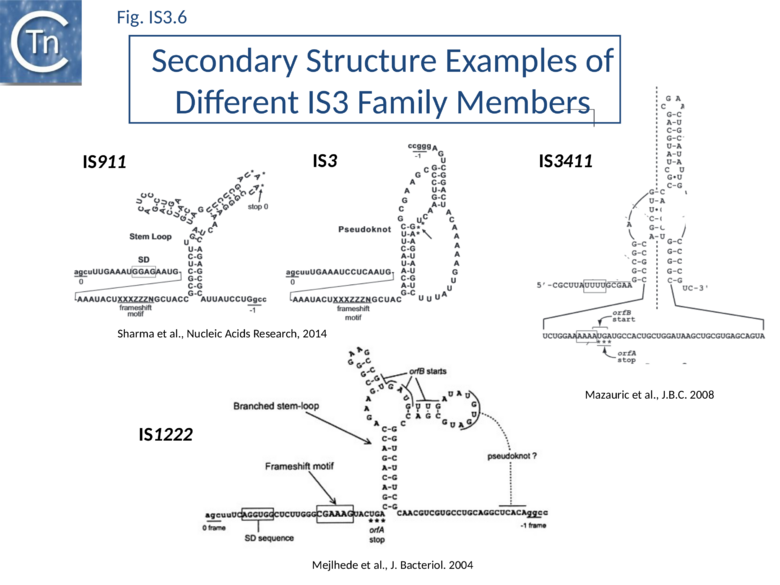
Two similarly located partially overlapping reading frames in IS3, IS150 and IS3411[54] also produce three proteins. The transposases, OrfAB, like that of IS911, are fusion products of the two orfs generated by a –1 translational frameshift.
For IS3, frameshifting is also stimulated by a presumed H-type pseudoknot structure similar to those generally involved in viral recoding[63]. In IS3411, -1 slippage on a U UUU motif requires a more convoluted form of pseudoknot structures formed by pairing of an apical loop and an internal loop belonging to two hairpins located 65 nucleotides apart on the mRNA[54]. Two similarly arranged orfs occur in IS2 and have been shown to encode OrfA and OrfAB equivalents only[26][56]. This organization is observed in most members of the IS3 family but, beside the cases mentioned above, frameshifting has been analyzed experimentally only in a few other, less well-characterized, elements (including IS51, IS222, IS600, IS1133, IS1222).
The frequency of frameshifting is quite variable from element to element: reported values are 15% for IS911, 50% for IS150, 6% for IS3 and 2% for IS3411[54]. These values may not reflect the in vivo situation since they were not established by direct measurement of the amount of the OrfA and OrfAB proteins synthesized from an intact IS, but after modification of expression signals of the IS genes or after cloning the frameshift signals in a reporter system[19][20][21].
The level of formation of a circular IS911 transposition intermediate IS911 carrying abutted left and right ends to generate an IRR-IRL junction (Transposition Pathway) measured by PCR indeed depends on frameshifting frequency in vivo[64]. IS911 copies from several clinical isolates contained variations in the frameshift region exhibited various reduced levels of frameshifting. When these were introduced into the model IS911 they resulted in comparable reductions in a circle formation.
Frameshifting is likely modulated by the physiological state of the host cells and by the environment: for example, frameshifting decreases when the temperature is raised or when ribosome density on the mRNA is increased (O. Fayet, pers. Comm.).
Artificial orfA-orfB fusion
For experimental purposes, production of OrfAB without necessitating a translational frameshift is obtained by introduction of a single additional base pair within the frameshift region which artificially fuses the orfA and orfB frames and eliminates OrfA production[21]. It was initially difficult to construct this mutant in the context of an entire IS911 (i.e. with the two flanking IR) but more recently this has been accomplished using a longer artificial IS and resulted in an exceptionally high transposition frequency[65]. A similar mutant in IS3 results in a high frequency of adjacent deletions[20].
Structural motifs
Although no structural information is available from crystallography, the role of the HTH and LZ motifs have been probed in vivo and in vitro.
The conserved N-terminal helix-turn-helix (HTH) motif is related to the LysR family of bacterial transcription factors and has a highly conserved tryptophan residue similar to that of certain homeodomain protein HTH motifs. This domain is important in directing transposase to bind IS911 IR[49] and is present in most IS3 family members (Fig.IS3.7 A). The N-terminal helices of the related IS2 transposase are also involved in IR binding[49].
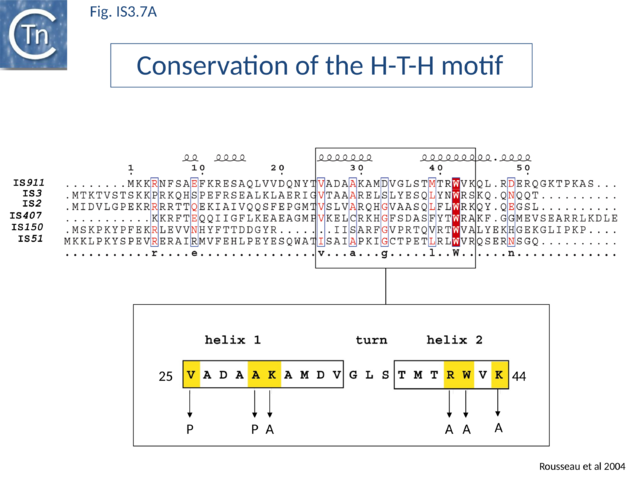
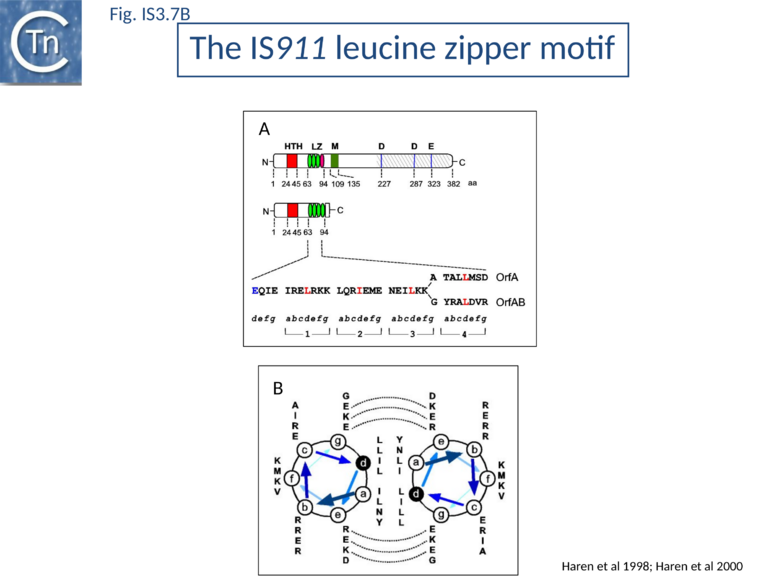
Many members carry a putative leucine zipper located at the end of OrfA (sometimes extending into the OrfB region of the OrfAB protein) (see [40] [66][67]). Studies with IS911 and IS2 indicate that this is a multimerization domain of the proteins[38][50][68]. The LZ motif of IS911 is composed of four heptameric units (Fig.IS3.1 B) with a predicted coiled coil structure including a potential buried inter-subunit hydrogen bond across the dimer interface (Fig.IS3.7 B), to maintain the zipper in a dimeric state, and correctly placed residues with opposite charges potentially able to form characteristic inter-subunit salt-bridges to stabilize the dimeric structure[50]. Leucine zipper motif are found in most IS3 family members (Fig.IS3.7 C).
OrfAB and OrfA form both homomultimers and mixed OrfAB-OrfA multimers[38][50].
Mutation of specific critical residues in the OrfAB LZ reduces the level of transposition intermediates in vivo and in vitro [69] (Transposition Cycle) and reduced or prevented multimer (dimer) formation. OrfAB and OrfA share three of their four heptads (Fig.IS3.7 B). The last of each differs in sequence due to the translational frameshift which occurs within the heptad in the expression of OrfAB. This presumably results in different strengths of monomer-monomer interactions in the case of homo- and hetero-multimers and this may be involved in the regulation of transposition. A poorly defined region, M, located between residues 109 and 135 (Fig.IS3.1 B) and components in the catalytic domain of OrfAB are also involved in its multimerization.
Co-translational DNA binding
IS911 OrfAB has a strong cis preference in vivo [65]. It has about a 200 fold higher activity on the IS copy from which it is expressed (in cis) than in trans. This prevents activation of transposition of one IS copy by OrfAB expressed from a second copy in the same cell. The strength of the cis effect depends on the distance of the transposase gene from the IS ends. Also, modification of the translational frameshifting pause signal has a strong influence on cis preference presumably by delaying translation and folding of the C-ter domain increasing the chance that the folded N-ter domain will recognize and bind its target IR.
In vitro analyses using ribosome display with a coupled E.coli-derived transcription-translation system coupled with size exclusion chromatography[65] demonstrated that an added IR bound nascent OrfAB derivatives while they are still attached to the ribosome. Ternary complexes containing mRNA, ribosome, and a nascent peptide specifically bound added IR copies if only the N-ter 149 amino acids extended from the ribosome whereas a full-length Tpase exiting the ribosome did not.
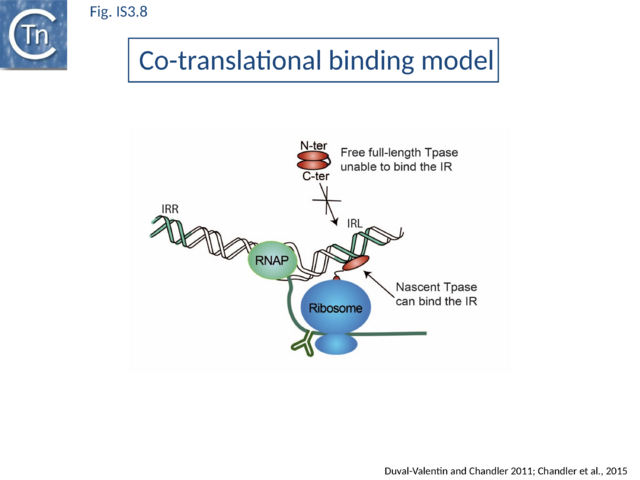
Direct evidence of coupled translational binding (Fig.IS3.8) was obtained using a staged coupled transcription/translation reaction: nascent OrfAB bound the IR before its synthesis was complete but not after. Thus OrfAB can efficiently bind the IR only prior to its complete translation.
Co-translational multimerisation
An intriguing question arising directly from these results is how OrfAB multimerizes as is found in the transpososome to bind both ends of the IS. Stable formation of the important synaptic complex containing both IS ends and the transposase requires a dimeric OrfAB (see "The IS911 transpososome" below). It is therefore possible that dimerization is in some way directly associated with translation. Indeed, using luxA and luxB as a model system, it been shown that luxA/B subunit assembly initiates cotranslationally on nascent LuxB in vivo. Protein assembly appears to be directly coupled to translation and involves “spatially confined, actively chaperoned cotranslational subunit interactions”[70].
The IS911 transpososome
A crucial checkpoint in transposition is the assembly of the 'transpososome'. This step is a general prerequisite for initiating DNA cleavage and the subsequent chemical steps in transposition for most elements that use a DNA (rather than RNA) transposition intermediates. In this protein-DNA complex, both ends of the transposon are bridged by the transposase before it catalyzes the DNA strand cleavages and strand transfers necessary for transposon mobility[71][72][73]. The transpososome adopts very precise architectures to accomplish these steps, and undergoes defined changes throughout the transposition process.
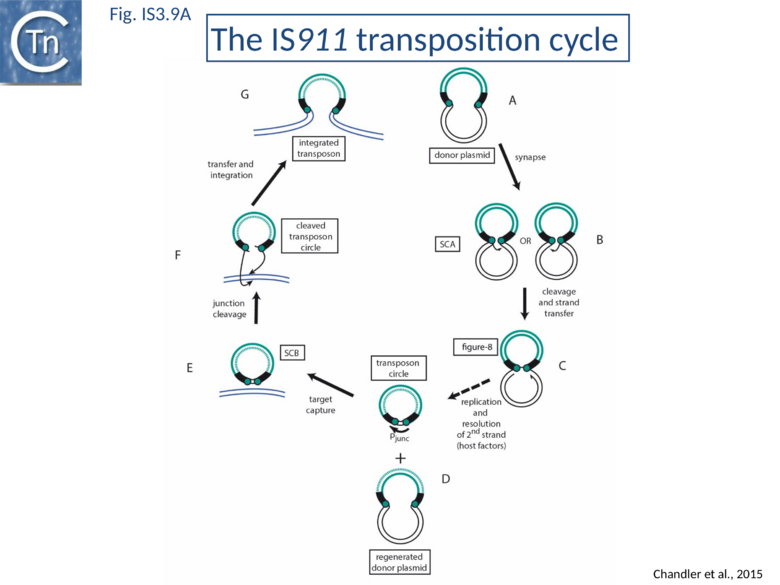
The overall IS911 transposition pathway is a two-step process, involving replicative excision followed by insertion (Fig.IS3.9 A and 9B). This implies consecutive assembly of two types of transpososome: one implicated in IS excision (synaptic complex A; SCA) and includes both IS ends while the other (synaptic complex B; SCB) involves the circle junction with its abutted IRs to ensure its integration into the target DNA.
Excision synaptic complex SCA.
Using a band shift assay and IR of different lengths (the so-called “long-short” experiment) it was shown that the truncated OrfAB [1-149] forms a complex with two IR copies, the paired-end complex (PEC)[38] equivalent to the SCA. An intact OrfAB [1-149] LZ is necessary for correct PEC/SCA formation[38][50]. At higher OrfAB [1-149] concentrations a probable single end complex (SEC) composed of one IR and OrfAB [1-149] appeared. Addition of OrfA disturbed both PEC/SCA and SEC and generated a fast migrating species whose composition remains to be determined but does not appear to contain OrfA itself [38].
DNaseI and Copper phenanthroline footprinting revealed that OrfAB [1-149] protects a sub-terminal (internal) IR region including two conserved sequence blocks in the left (IRL) and right (IRR) ends (Fig.IS3.1 A). DNA binding assays in vitro and measurement of in vivo recombination activity of sequential IR deletion derivatives suggested a model in which the N-terminal region of OrfAB binds the conserved boxes in a sequence-specific manner and anchors the two IRs into the SCA. The external region of the inverted repeat was proposed to contact the C-terminal transposase domain carrying the catalytic site[74].
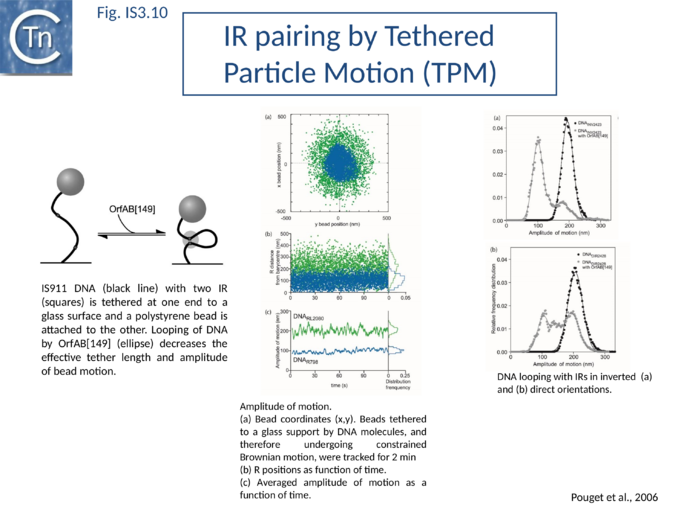
SCA is composed of a dimer of transposase bridging to two IR[75], as judged by the use of a tagged and untagged truncated transposase derivative, OrfAB[1-149], and also of IR of different lengths. OrfAB[1-149] assembles two IRR copies in a parallel orientation (Fig.IS3.4)[75] as studied at the single molecule level by Atomic Force Microscopy (AFM) using asymmetric IRR-carrying DNA fragments.
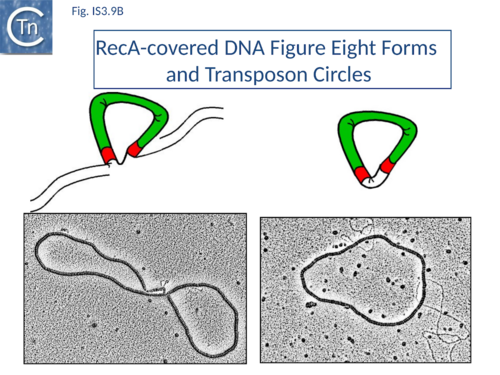
SCA assembly was also studied using a second single-molecule approach: tethered particle motion (TPM) (Fig.IS3.10)[76] in which a DNA molecule is tethered to a glass support and its effective length is measured by observing the Brownian motion of a bead attached to its free end (Fig.IS3.10 left). OrfAB[1-149] binding to a single IR provoked a small shortening of the DNA, consistent with a DNA bend introduced by protein binding to the IR and was confirmed using EMSA. When two ends were present on the tethered DNA in their natural, inverted, configuration, OrfAB[149] not only provoked the short reduction in length but also generated species with greatly reduced effective length (Fig.IS3.10 middle and top right) consistent with DNA looping between the ends and thus SCA formation. SCA is very stable and kinetic analysis in real-time suggested that passage from the bound unlooped to the looped state could involve another unlooped species of intermediate length in which OrfAB[149] is bound to both IRs. DNA carrying directly repeated IR also gave rise to the looped species but the level of the intermediate species was significantly enhanced (Fig.IS3.10 middle and bottom right). Its accumulation could reflect a less favorable SCA formation with directly repeated IR copies than with inverted IR. This is compatible with a model in which OrfAB binds separately to and bends each IR and protein-protein interactions then lead to SCA formation (Fig.IS3.11 A)[77]. Cleavage and strand transfer would then give rise to a species in which both IS ends are joined by a single strand bridge (or figure-eight on a circular plasmid (Fig.IS3.9 C) (see "The Transposition Pathway").
Insertion synaptic complex SCB
SCB has not been characterized in such a precise way as SCA. SCB is devoted to the insertion step of the transposition process. Two types of insertion, IR-targeted and non-targeted, have been observed (Fig.IS3.11 B). It has been proposed that two different protein-DNA complexes are assembled during the two types of insertion reaction: SCBt and SCBnt (for targeted and non-targeted synaptic complex respectively[78]. Nothing is known about the stoichiometry and the geometry of these complexes but, based on protein and DNA requirements for protein-DNA complex formation, as judged by band shift, and for transposition products, as judged by in vitro and in vivo transposition assays, it has been proposed that SCBt is composed of a transposase dimer bridging a DNA molecule carrying an IR and a DNA molecule carrying an IRR-IRR junction (IS911 circle), the product of the replicative IS911 excision. This IR targeted insertion explains how the original isolate of IS911 might have occurred next to a sequence which strongly resembles an IR[11] and can also explain one ended insertion[33]. In this regard, IRR shows a somewhat higher affinity than IRL. Note that if one of the two IR carried by the circle is omitted, SCBt resembles SCA (Fig.IS3.11).
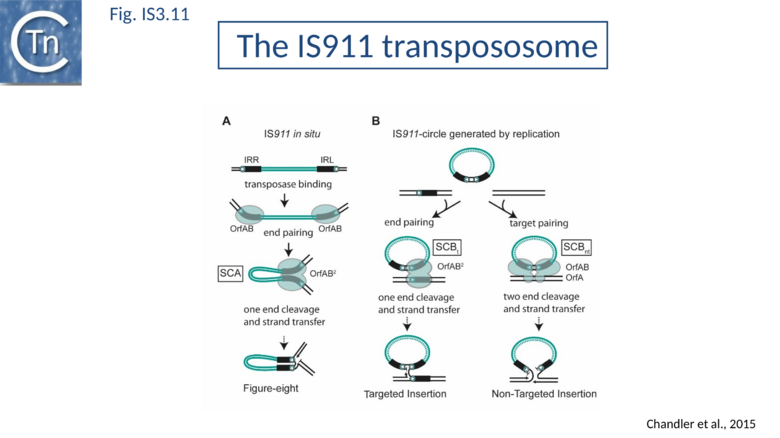
SCBnt is thought to differ from both SCA and SCBt and to include the second IS911 protein, OrfA. This protein, binds non-specifically to DNA and interacts with OrfAB[38][50], is proposed to direct an OrfAB-junction complex to a randomly chosen target-DNA to form SCBnt[78][79]. This is based on the observation that integration of the transposon circle intermediate is greatly stimulated by preincubation of OrfAB and OrfA in an in vitro reaction[80].
The Transposition Pathway
The IS3 family is one of an increasing number of IS families known to transpose using a double strand circular DNA intermediate. Closely related pathways have been demonstrated for IS1[81], IS2[22], IS3[82], and IS150[83]. This represents a major transposition pathway which has yet to be widely recognized. As shown in Fig.IS3.9, and the animation below, IS3 family transposition proceeds through a copy-out-paste-in process.
IS911. copy-out-paste-in mechanism
|
|---|
The Figure-eight form
The initial step is recognition of the IR by OrfAB (presumably during its translation) (IS911 movie above) and assembly of SCA to correctly position the DNA ends and the transposase catalytic site for the subsequent chemical steps. Like all known DDE transposase-catalyzed reactions[84], IS911 transposition proceeds by cleavage of a single strand at the transposon end generating a 3’-OH. This then attacks a target phosphodiester bond in a strand transfer reaction. The particularity of this copy-out-paste-in mechanism is that initial cleavage occurs at only one transposon end, either left or right (Fig.IS3.9). This single liberated 3’-OH directs strand transfer to the same strand 3 bases 5’ to the other end of the element. This generates a molecule in which a single transposon strand is circularized to produce a single strand bridge generating a figure-eight structure on a circular plasmid donor molecule (Fig.IS3.12) which can be easily observed in vivo[85]. The IRs are joined by the single-stranded bridge and separated by three bases derived from flanking DNA from either the left or right end. The three (or 4) bp direct repeats flanking the original insertion are not required for further transposition (as also shown for IS3[86]) and an IS911-based transposon engineered to have different flanks generates a mixed population of figure-eight molecules with one or other flank sequence. Prevention of cleavage of one or other transposon end resulted in a homogenous population that carries the 3nt DNA flank associated with the mutant end confirming that the IRL can attack IRR and vice versa. The reaction can be viewed as a one-ended site-specific transposition event. These initial steps can be accomplished by OrfAB alone. However, it should be noted that in the presence of OrfA, no figure eight or IS circles could be detected by a simple gel assay in vivo although IS circles were found using a PCR approach[64]. This suggests that OrfA may play a role in negatively regulating the initiation of transposition. A similar conclusion has been reached for OrfA of IS3[87]. Alternatively, OrfA may stimulate the disappearance of figure eight and IS circles (see below) since no effect of OrfA was observed on figure-eight formation in vitro. Together with the fact that OrfAB is normally produced at low levels from a weak promoter[21], initiation of transposition to form the figure eight intermediate may be stochastic.
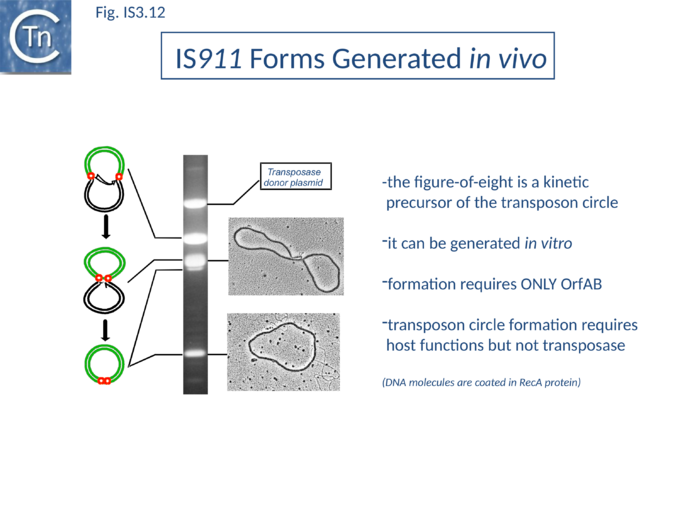
The circular intermediate
Kinetic data[65][85] indicate that the figure-eight gives rise to the circular transposon form which can easily be detected in vivo and in which the IR are abutted and separated by three base pairs of DNA flanking the original insertion (Fig.IS3.9 and Fig.IS3.12). As for figure-eight molecules, a transposon engineered to have different flanks generates a mixed population of transposon circles with one or the other 3bp flank located at the junction[88].
Studies in vivo using a labeling protocol and a temperature-sensitive plasmid as transposon donor demonstrated that conversion from the figure-eight to the transposon circle occurs by semiconservative replication where the circular intermediate is “copied out” leaving a copy in the transposon donor molecule[89] (Fig.IS3.9). This is transposon-specific, requires OrfAB (presumably to generate the figure eight and generate a 3’-OH on the IS911 DNA flank) and does not depend on replication from the donor plasmid origin of replication[89].
Using donor plasmids where one or other IR was inactivated for cleavage would be expected to determine whether one or other of the 3’-OH is used in transposon replication. This was tested using the Tus/ter system[90][91][92][93] (which blocks passage of a replication fork in an orientation specific fashion) cloned into the transposon in either one or other orientation. In the presence of Tus protein, no transposon circles were observed if the orientation of the ter site was that expected to block replication from one or the other end[89].
At present, it is not known how OrfAB is removed and how this replication step is initiated or terminated to generate the final circles. It is possible that these processes involve host factors and mechanisms similar to those, which operate in replicative transposition of bacteriophage Mu (see [94][95][96]).
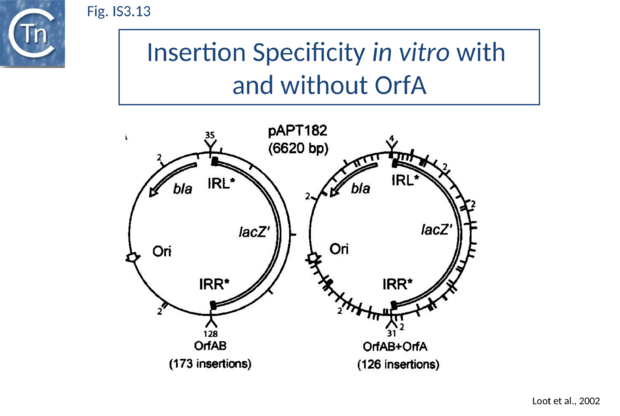
RecG helicase is implicated in targeted insertion. This process involves a target IS911 end and strand transfer occur between one cleaved end of the IS circle and the target IS end to create an intermolecular single-strand bridge rather than the intramolecular bridge of the figure-eight intermediate (Fig.IS3.13). Resolution of this structure implicates branch migration and replication from the donor plasmid[97]. This reinforces the idea that host proteins including components of the replication machinery are loaded onto figure-eight intermediates.
Integration of the circular intermediate
The IR junction formed by IS circularization is very unstable in the presence of OrfAB and undergoes high levels of deletion and insertion in vivo[98] and in vitro[80]. Transposon circle insertion presumably requires further transposase synthesis.
A remarkable consequence of transposon circle formation is the assembly of a strong promoter, pjunc, from a –35 hexamer contributed by IRR and a –10 hexamer contributed by IRL (Fig.IS3.3 B). The 3 (or more rarely 4) bp which separate IRL and IRR in the circle provide an ideal spacing between the –35 and –10 elements[98]. The junction promoter, pjunc, is 30-50 fold stronger than the indigenous promoter, pIRL[98] (Fig.IS3.4), and more than two fold stronger than lacUV5[30]. It is correctly placed to drive high levels of transposase synthesis and plays an active role in controlling IS911 transposition.
Inactivation of pjunc by mutagenesis strongly reduced IS911 transposition in vivo when transposase was expressed in its native configuration[30]. Moreover, the truncated OrfAB derivative, OrfAB[1-149] , which specifically binds IRR and IRL, reduced in vivo promoter activity 10 fold in a mutated junction resistant to cleavage. Full-length OrfAB, which binds the IR only weakly, and OrfA, which does not specifically bind the IR, had no effect[30]. Integration results in disassembly of pjunc providing a powerful feedback mechanism resulting in transient and controlled activation of integration only in the presence of the correct (circular) intermediate.
For the related IS2, this junction promoter is required for transposition[99].
Circle junction formation brings both transposons ends together in an inverted orientation. This active junction must then participate in the second type of synaptic complex which includes target DNA (Fig.IS3.9 and Fig.IS3.11 B).
Two single strand cleavages, one at each abutted IR, would linearize the transposon circle permitting the two liberated 3'-OH groups to direct coordinated strand transfer (Fig.IS3.9 and Fig.IS3.11 B). The final step requires OrfAB but is greatly stimulated by OrfA and is sensitive to the ratio of OrfAB/OrfA[80].
It is not known whether target capture occurs before or after cleavage of the circle junction although it has been observed that linear copies of IS911 are produced from transposon circles in vitro and in the presence of high OrfAB levels in vivo and a pre-cleaved linear transposon was a robust substrate for integration in vitro[100].
Based on kinetics and on the formation of the strong pjunc promoter, we favor a model in which the IS circles represent a reservoir of transposition intermediates and that linear forms are generated from the IS circles during the integration process.
This has also been proposed for IS3[86].
Targeted Insertion
As stated above, several IS including IS911 show a preference for integration next to sequences in the target similar to their IR. One way of understanding this is that the transposon circle is able to form a synaptic complex (SCBt; Fig.IS3.11 B left) which is similar to SCA (Fig.IS3.11 A) but which occurs “in trans” between an IR of the transposon circle and an IR in the target. In the case of IS911, this phenomenon occurs more frequently if OrfA is not present (Fig.IS3.13) and it was proposed that one role of OrfA is to promote dispersion of the IS[78][101].
This type of one-ended intermolecular recombination/integration has been analyzed in some detail[97][101][102].
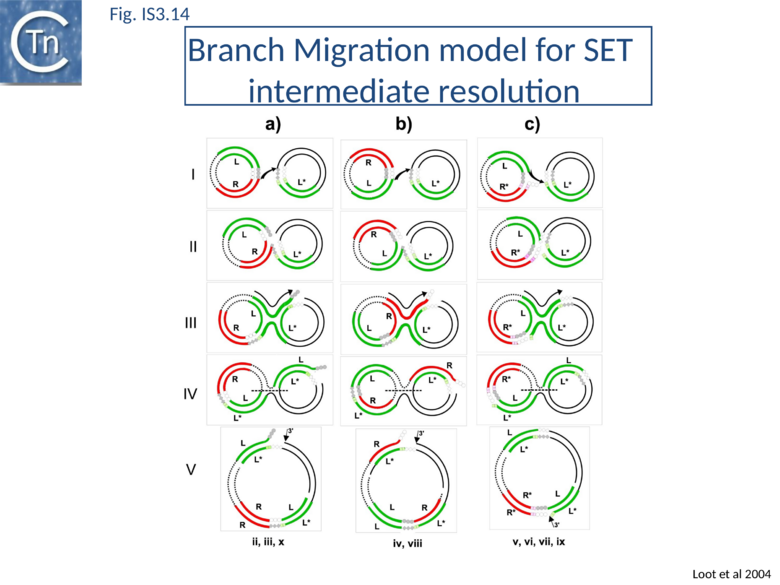
IR-targeted insertion involves the transfer of a single end of the junction to the target IR to generate a branched DNA structure. The single-end transfer (SET) intermediate, but not the final insertion product, was detected in vitro. This implies that SET intermediates must be processed by the bacterial host to obtain the final insertion products. Sequence analysis of in vitro and in vivo IR-targeted insertion products revealed high levels of DNA sequence conversion in which mutations from one IR were transferred to another. These sequence changes could not be explained by the classic transposition pathway but could be understood in terms of a mechanism in which SET generates a four-way Holliday-like junction which is then processed by host-mediated branch migration, resolution, repair and replication. This pathway resembles those described for processing other branched DNA structures such as stalled replication forks. A version of this model is shown in Fig.IS3.14. Subsequent studies showed that the RecG helicase is implicated in vivo, as might be expected for strand migration[97].
Mechanism in other family members
Several other members of this family have also been analysed in some detail. These include IS2, IS3, and IS150. All three have been shown to generate circles when supplied with high levels of the fused frame Tpase[20][22][83][86][103].
IS3 also generates adjacent deletions[20] but, unlike IS911, appears to undergo excision from the donor molecule as a linear form following a staggered double strand break at each end. These forms have a 3 base 5' overhang and may be an alternative type of transposition intermediate[103]. Such forms may be equivalent to the linear IS911 species derived from transposon circles. In addition, IS3-derivative transposons in which two abutted ends have been engineered undergo high levels of transposition[31].
Insertion of IS3 creates generally 3 and sometimes 4 bp direct target repeats. It is significant that plasmids in which the IRs are separated by 4 bp are more active than those separated by 8 bp. In these studies, the authors were unable to engineer derivatives with two complete tandem IS3 elements. This may be the result of the formation of a strong hybrid promoter which, as described for IS911 and other ISs (see above), drives high levels of Tpase expression. This configuration of ends is equivalent to that found at the circle junction and suggests that abutted ends of IS3 are also efficient substrates in transposition.
IS2 generates direct target duplications of 5 bp on insertion[104] although transposon circles generated with this element carry only a single base pair separating IRL and IRR[22].
While IS2 carries a conserved terminal 5' -CA- 3' at its right end, the left end terminates with 5' -TG- 3'. This atypical IRL does not act as a strand donor but uniquely as a target in the circularization reaction.
Functional studies indicate that the product of the upstream orfA may inhibit transposition[26]. It has been shown to bind specifically to IRL at a sequence that overlaps the -10 hexamer of the resident Tpase promoter and represses expression of OrfA.
It does not appear to bind IRR (note that in the original article the authors inverse the standard definition of IRL and IRR[26].
Several other elements also exhibit small inverted repeat sequences which flank the -10 hexamer of the putative resident Tpase promoter. IS2-derivative transposons in which two abutted ends have been engineered also undergo high levels of transposition[22][105] and, like IS911, the circle junction of IS2 also constitutes a strong promoter capable of driving Tpase expression. Several (but not all) IS3-family elements may also carry similarly located potential -35 and -10 sequences within their IRs.
Structural studies
Although there are at present no structural data available for any members of this family, recent results obtained with an IS from another family, ISCth4 from the IS256 family, which also undergoes copy-out-paste-in transposition has provided some insights [106]. This particular transposition pathway is asymmetric in the sense that one IS end is cleaved and attacks the opposite end several nucleotides from the tip [85]. In accord with this type of mechanism, crystal structures of ISCth4 transposase bound to three different substrates show a transposase dimer bound asymmetrically to a single DNA substrate: a pre-reaction substrate with IRR together with its flanking DNA, a pre-cleaved complex in which the IRR flank had been removed and a strand transfer complex including an abutted IRR and IRL separated by a gapped 6 base pair linker (Fig. IS256.8).
It is important to note that IS256 family transposases carry an alpha-helical insertion domain which separates the catalytic domain into two segments. This domain plays an important role in directing different DNA segments during the reaction. IS3 family transposases carry an uninterrupted catalytic domain without the alpha helical insertion domain implying that the atomic details of the process will differ. In this light, it is worth remembering that efficient insertion of IS911 transposon circles catalysed by OrfAB is greatly stimulated by inclusion of the upstream OrfA protein and is sensitive to the ratio of OrfAB/OrfA [107].
Excision: A dedicated enzyme
This section has been published in a modified form as Chandler M, Ross K, Varani AM. The insertion sequence excision enhancer: A PrimPol-based primer invasion system for immobilizing transposon-transmitted antibiotic resistance genes. Mol Microbiol. 2023 [108].
The IS3 family insertion sequence IS1203v (similar to IS629), originally identified in a Shiga toxin 2 gene (stx2) of Escherichia coli O157:H7 which it had insertionally inactivated, was found to undergo precise excision leading to stx2 reactivation [109]. Curiously excision of the IS3 family transposon, IS1203v occurred at a much higher frequency in some in some E.coli hosts than in others [110].
IS Excision is Stimulated by High Transposase Levels
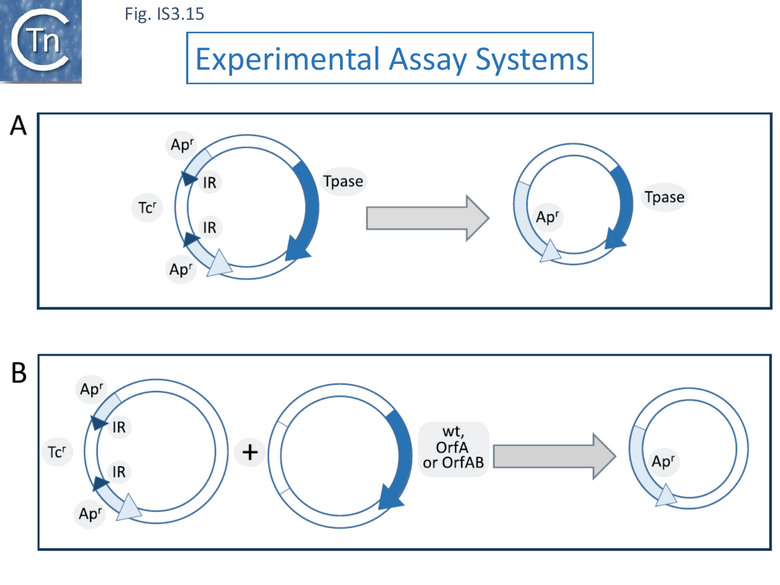
Using a (single copy) F plasmid derivative in which an ampicillin resistance gene was interrupted by an IS1203v insertion (bla::IS1203v) to monitor precise excision rates (reversion to ampicillin resistance) (Fig. IS3.15 A), the authors showed that excision was 105 fold higher in Escherichia coli O157:H7, known to carry a significant number of IS629 copies, compared to E. coli K12 (MG1655), where it is absent [111][112].
Further studies using a number of E. coli isolates with and without IS1203v/IS629 copies supported the idea that excision was higher in those strains already carrying the IS.
In a modified experimental system in which various transposition functions were supplied in trans from a compatible plasmid (Fig. IS3.15 B), deletion was observed to be very low or below the detection level in an MG1655 host. In the O157:H7 strain, however, supplying the OrfAB transposase induced high deletion levels (~10-3) compared to that obtained with the empty vector (7.8x10-7) whereas supplying the orfA, orfB and orfAB genes in their native configuration only resulted in a moderate frequency of excision (2.6x10-6) and supplying orfA alone depressed excision (3.8x10-9). This implies that the levels of available OrfAB transposase are a determining factor in excision. A survey of a number of strains showed that excision frequencies of strains possessing IS1203v (IS629) were on average 103 times higher than those not carrying the IS. High IS629 excision frequencies were observed in a large number of clinical E. coli isolates [113].
A Dedicated Enzyme: Identification of a common reading frame ECs1305 in all high excision strains
The authors identified a reading frame, ECs1305, present in all high excision strains that was absent in the low excision strains [115]. In EHEC O157 it It is located in large potential integrative elements that are similar to SpLE1 of EHEC O157 [114] and has probably been dispersed in this way. It was identified both in enterohemorrhagic (EHEC) and enterotoxigenic (ETEC) E. coli strains but homologues were also identified by Blast analysis in a broad range of bacteria including Alpha- Beta-, Gamma-, Delta- and Epsilon-proteobacteria; Bacteroides; Chlorobi; Cyanobacteria; Firmicutes; Actinobacteria; and Verrucomicrobia[115].
More recently it has been estimated that a highly conserved IEE gene copy is present in over 30% of available E. coli genome assemblies and is very abundant not only within enterohemorrhagic and enterotoxigenic genomes but also within enteropathogenic E. coli [116].
The ECs1305 gene was subsequently named iee for IS-excision enhancer [115]. In EHEC O157, ECs1305/IEE is located in a large potential integrative element that is similar to SpLE1 and has probably been dispersed in this way [114].
ECs1305 and an active transposase is required for high level IS excision
When this reading frame was deleted, the IS excision frequency was greatly decreased but could be restored by reintroduction of a plasmid-carried iee copy. Moreover, the use of DDE mutant transposases was used to demonstrate that the ECs1305-promoted excision behavior was also dependent on an active transposase.
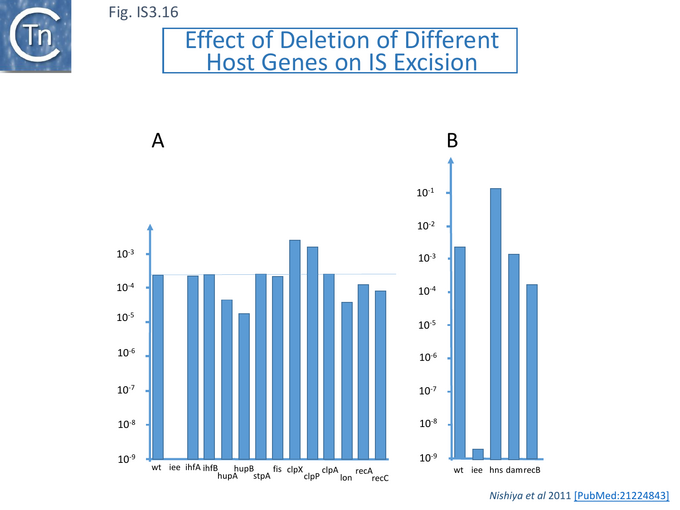
The effect on excision frequency of a number of host genes was investigated and although some of these had been shown to affect excision of other ISs (e.g. [117][118][119] ), the effect of mutations were largely marginal (Fig.IS3.16) and not as pronounced as mutations in ECs1305 [115].
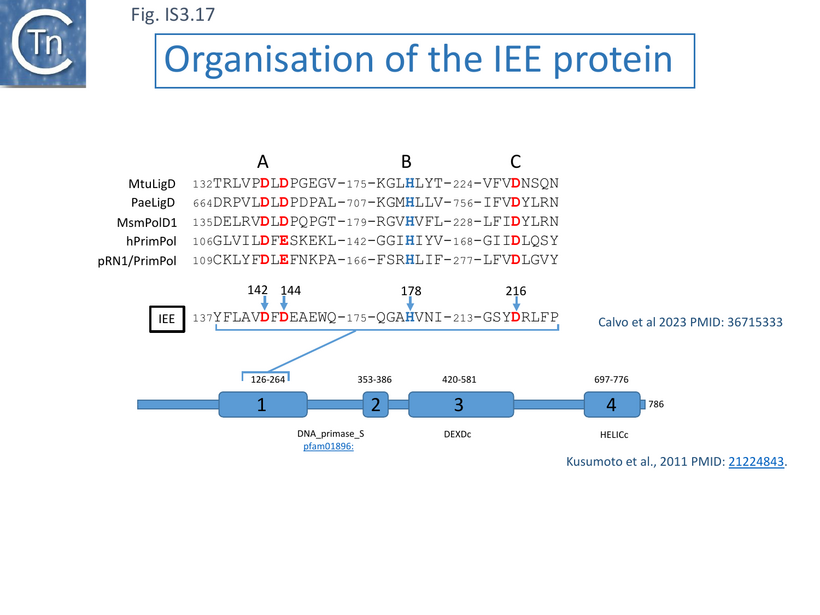
Analysis of the primary Sequence of IEE: a PrimPol Helicase
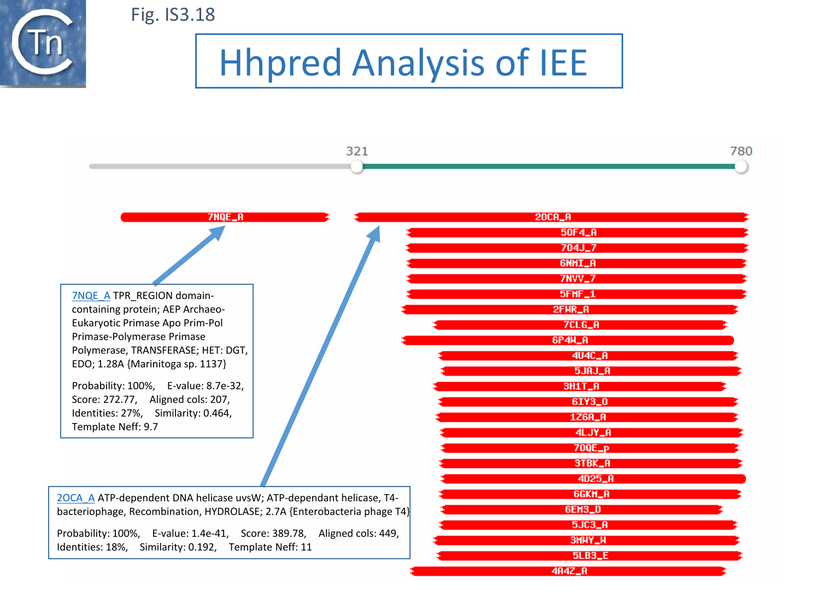
Alignment of a number of IEE proteins revealed 4 conserved regions [115]. Two of the more N-terminal regions (1 and 2) showed regions with similarities to eukaryotic/archaeal/bacterial primase domains (AEP) while the more C-terminal regions 3 and 4 showed similarities with DEAD and DEAH box helicases [115] (Fig. IS3.17). Analysis using hhpred (https://toolkit.tuebingen.mpg.de/tools/hhpred/ ) indicates that the N-terminal domain resembles the AEP Archaeo-Eukaryotic Prim-Pol Primase-Polymerase (Fig. IS3.18) while the C-terminal domain is similar to the ATP-dependent DNA helicase, UvsW, of bacteriophage T4. The authors find that targeted mutagenesis of the helicase domains “considerably reduces IEE activity” [115].
Biochemical Analysis of IEE Activities: Polymerase Activity
Indeed, as proposed from alignments with other members of this protein family [115][116] (Fig. IS3.17), biochemical analyses using purified IEE demonstrated that the enzyme possesses DNA polymerase activity in the presence of Mg2+ [116] and can extend a 15 bp primer on a short 33bp template using dNTPs but not NTPs (Fig. IS3. 19 i). Mutation of two of the probable AEP catalytic residues (Fig. IS3.17), D142A/D144A, eliminated this activity.
As might be expected from other examples of a number of endonucleases and nucleotidyl-transferases, the polymerization reaction was significantly higher in the presence of Mn2+ ions and the extension length was increased.
Experiments using various molar ratios of Mn2+ and Mg2+ led the authors speculate that the IEE AEP domain might preferentially use Mn2+ in vivo even in the presence of Mg2+. They argue that the active site of a number of AEP is more flexible than that of replicative polymerases since is configured to “accommodate dislocations of the template and primer strands as well as to extend mismatched base pairs” [116]. These include the AEP domains of the Translesion Synthesis, TLS, class such as human PrimPol, and those of the Non Homologous End Joining, NHEJ, class such as bacterial LigD. They suggest that this flexibility allows preferential accommodation of Mn2+.
Preferential IEE use of Mn2+ suggested that it too could also allow extension with mismatched base pairs and promote DNA strand rearrangements during polymerization. This was demonstrated using a set of substrates carrying a single or double base changes in the template strand (Fig. IS3.19 ii and 19 iii) while supplying individual defined nucleotides in the reaction. Although the enzyme showed a strong preference for incorporation of complementary nucleotides at the templating nucleotides (Fig.IS3.19 ii) during primer extension, a significant degree of misincorporation (~16%) also occurred [116].
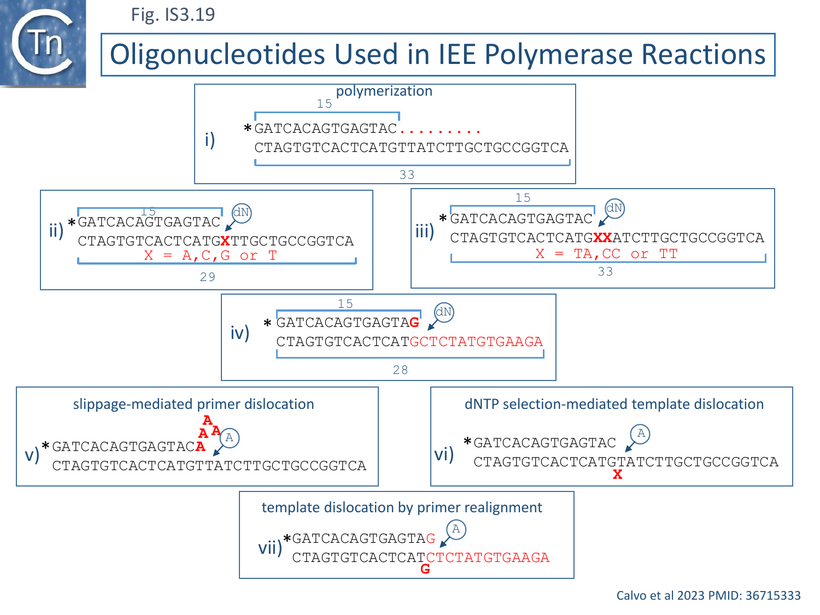
Interestingly, a major product resulted from insertion of two (identical) nucleotides on substrates in which the first and second templating nucleotides were different. In cases where these were the same, addition of the complementary nucleotide to the reaction resulted in an “expansion” by “reiterative primer strand slippage to extend it by 5-7 nucleotides as shown in Fig.IS3.19 v. This was observed for both the TT and CC template pair where reiterative addition of A or G occurred respectively. IEE could also bypass the first template base in substrates as shown in Fig.IS3.19 v and bypass a mismatch (Fig. IS3.19 iv) as schematized in Fig. IS3.19 vii.
The authors conclude that their results indicate that IEE is an “error-prone DNA polymerase” which can undergo slippage-mediated primer dislocation (Fig.IS3.18 v), dNTP selection-mediated template dislocation (Fig.IS3.19 vi) and template dislocation by primer realignment (Fig.IS3.19 vii), all resulting in DNA distortion. Dislocation of primer and template strands would, of course, facilitate the search for microhomologies.
Microhomology-Mediated End-Joining
These properties raised the question of whether, like bacterial end-joining PolDom, the primer/template dislocations IEE AEP domain could promote Microhomology-Mediated End Joining, MMEJ. This might explain how the enzyme stimulates IS excision using its reduced nucleotide insertion fidelity (as shown in Fig.IS3.19).
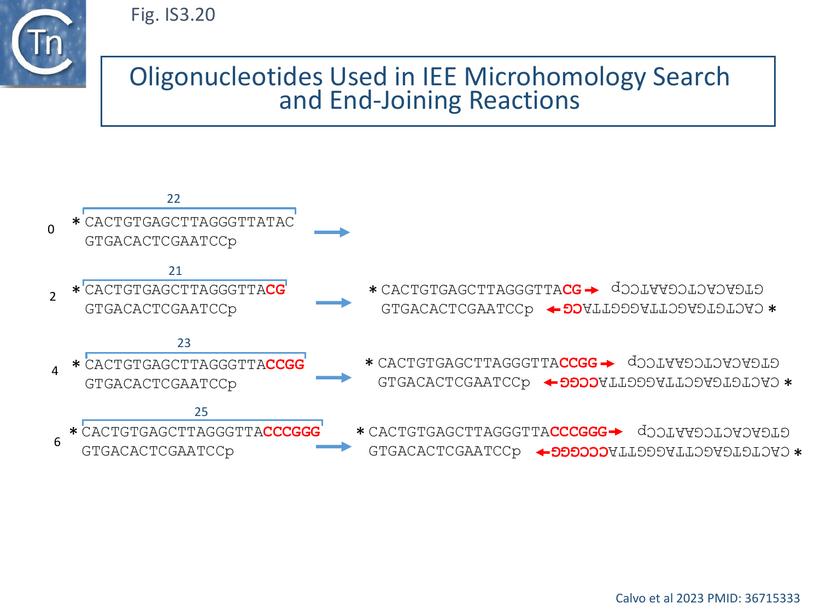
Using appropriately 5’ resected double strand oligonucleotides (Fig. IS3.20) with 0, 2, 4 and 6 nucleotide microhomologies at their 3’ ends (Fig. IS3.19 left). IEE- mediated MMEJ would create 3’ ends which should act as primers on 3’-synapsed DNA (Fig. IS3.20 right). A polymerization reaction including the four dNTPs revealed that IEE could specifically elongate a significant proportion of the 4 and 6 nucleotide substrates (28 and 46%) although no detectable elongation was observed with the 2 nt substrate under the reaction conditions used. IEE was also capable of limited synapsing of single strand substrates with 3’ terminal microhomologies [116].
The C-terminal Helicase Domain
IEE exhibits DNA-dependent ATPase activity which is eliminated in an active site K451A mutant. However, it was not able, as do other ATP dependent helicases, to couple its ATPase activity with unwinding of double strand DNA with 3’ or 5’ unpaired tails. A number of experiments comparing the activities of full length IEE and a C-terminal truncation which deletes the helicase domain from residue 289 (Fig. IS3.17) suggested that the C-terminal domain plays a role in stabilizing the interaction of the full length protein with its DNA substrates. The authors propose that it might even contribute to the removal of transposase from the transposition complex [116].
Models for IEE activity
Using a non-selective PCR approach, IEE-enhanced excision was shown not only to increase precise excision but also other, more extensive, deletion events. It was hypothesized that these occurred as a consequence of the IS3 family transposition mechanism in which initial cleavage of one IS end is followed by its strand transfer close to the opposite end to form a bridged molecule containing a small flanking sequence from the vector plasmid at the junction (Fig.IS3.21 i) (see IS3 The Transposition Pathway). The deletions were explained by the hypothesis that this initial strand transfer could be ‘sloppy” and target DNA sequences other than a second end (Fig.IS3.21 i).
However, transposition of this family of IS occurs by a copy-out mechanism and is thought to regenerate the donor plasmid (Fig.IS3.21 i). Such deletions would not be consistent with this mechanism but would require an additional type of reaction to resolve the deleted donor.
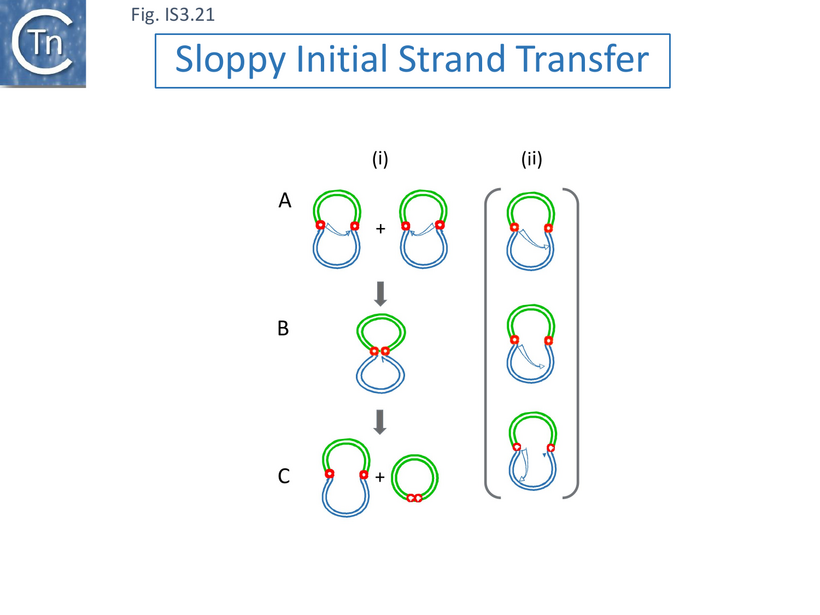
Calvo and coleagues [116] provide a model for IEE activity based on their biochemical results in which they propose that IEE pairs two 5’ resected ends DNA ends in a reaction which is facilitated by the C-terminal helicase domain, allowing the AEP domain to accomplish a “filling in” polymerization reaction. The model invokes an intermediate which is thought to occur during a cut-out-paste-in transposition pathway which leaves a blunt ended double strand break in the donor DNA molecule (see [120]). Moreover, although it is not pointed out, most IS generate short, direct target repeat sequences on insertion (see: General Information: What is an IS?) and would therefore provide 3’ terminal microhomologies upon 5’ resection.
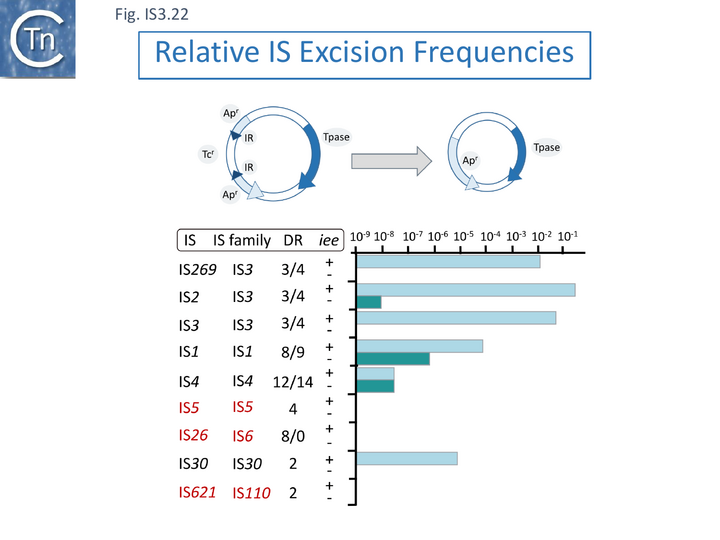
However, in addition to IS3 family members, Kusumoto et al., [115] show that IEE also stimulates excision of IS1 and IS30 family members (Fig. IS3.22), both of which can transpose using a copy-out-paste-in mechanism [81][121]. A number of the IS exhibit a measurable level of excision in the absence of the iee gene. Interestingly, it was proposed based on identification of in vivo transposase-induced structures, that IS1 can transpose using a number of alternative pathways [81]: IS1 shows a low basal level of excision which is further stimulated by IEE. It is possible that excision in the absence of iee occurs by a different mechanism [122][123] as occurs in the IS4 family member, IS10. In this light, it is noteworthy that IS4 itself shows a low level of iee independent excision which is not affected by iee (Fig. IS3.22). Additionally, IS5, IS26 and IS621, none of which use a copy-out-paste-in transposition mechanism, were not observed to excise even in the presence of iee.
Thus the common mechanistic property of those IS whose excision is stimulated by IEE is that they all use a copy-out pathway.
An alternative explanation to that of Kusumoto et al., [115], is that they are generated not during the first strand transfer step of copy-out-paste-in transposition as proposed (Fig. IS3.20), but during the second, replication step (copy-out) by slippage and realignment of the replication primer as was later proposed for the deletion of IS30 family ISApl1 copies flanking the colistin resistance, mcr, gene in Tn6330 [124] and Fig. IS30.10. This strand switching or primer invasion model had been proposed for the deletion of IS30 family ISApl1 copies flanking the colistin resistance, mcr, gene in Tn6330 [125].
The mcr connection: a model for excision by replication-associated strand exchange and primer invasion.
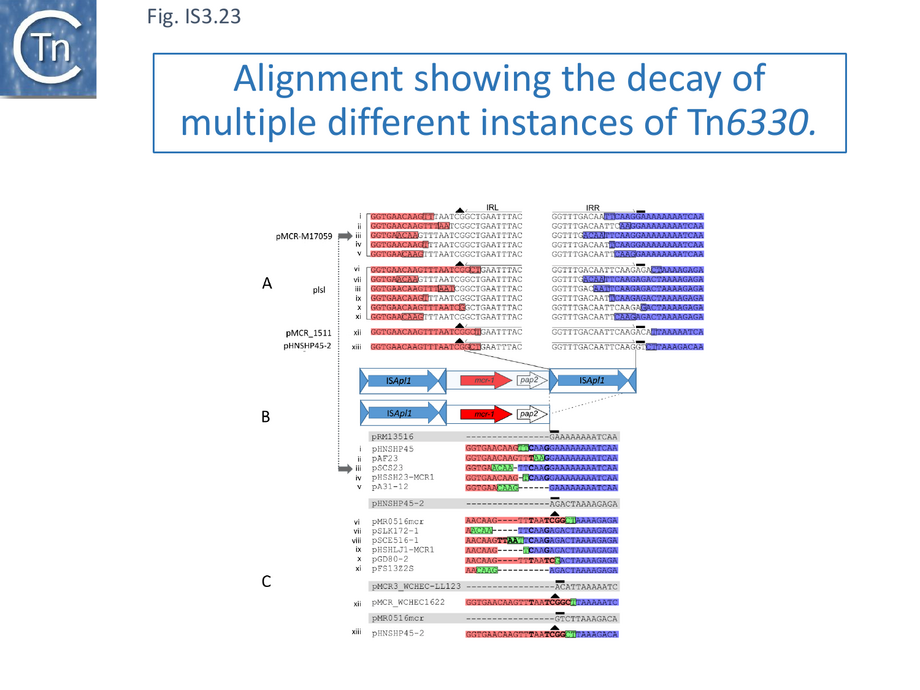
Colistin (polymixin E) is a last resort antibacterial that was used extensively in husbandry. Discovery of a transferable phosphethanolamine transferase conferring resistance to colistin in 2015 [126] was of such concern that it stimulated an immense effort to identify the resistance gene, mcr, in various bacterial sources worldwide. This quickly generated an extensive mcr sequence library in which it was noted that the gene was often, but not always, associated with an upstream or downstream copy of an IS30 family sequence, ISApl1. A number of examples carried two flanking IS copies, and the entire structure, which was proposed to be a compound transposon with characteristic 2bp direct target repeats [125], was called Tn6330 [127] and subsequently confirmed to undergo transposition [128]. Tn6330 carries the mcr1 gene together with a downstream open reading frame, pap2. Examination of a significant number of structures lacking the downstream IS revealed that the ~2.6 kb region including mcr-1 and pap2 was 99% identical; the non-identical nucleotides were concentrated at the 3’ end of pap2, the end that carries the downstream ISApl1 copy in Tn6330 (see Fig.IS3.22 B). Moreover, AT rich the pap2 gene in these cases was flanked by AT-rich regions. ISApl1 shows a strong preference for AT rich target sites, suggesting that an ancestral downstream ISApl1 copy had been deleted.
The number of available sequences was such that examples of closely related plasmid backbones could be identified which either had a complete Tn6330 insertion, with “empty” sites (often in multiple examples) as well as cases in which examples also inserted into the same location in which one or the other flanking IS was absent.
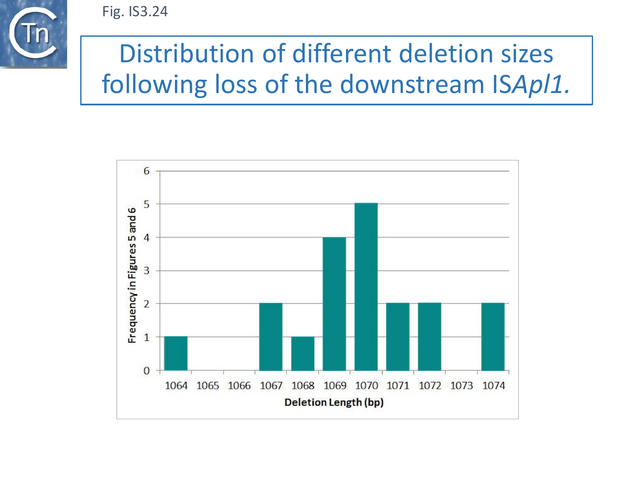
Careful scrutiny of the sequences flanking pap2 without an associated downstream IS, revealed small microhomologies which were thought to represent scars of deletions. Figure IS3.23 shows 4 plasmids each containing Tn6330 (pMCR-M15709, plsl, pMCR_1511, pHNSHP45-2; Fig.IS3.23 A). The salmon colored boxes on the left represent pap2 sequences retained in the subsequent deletions and the purple boxes on the right represent the external flanking sequences retained (some of these intrude into the IS). Figure IS3.23 C shows individual “deletants” with similar backbones to those shown in Figure IS3.23 A. It was noted that, when aligned, the deletions have largely occurred between microhomologies of two to four base pairs. Similar results were obtained when the sequences upstream of mcr1 were examined. Interestingly, the length of the deleted segments (Fig.IS3.24) remains close to that of ISApl1, 1070 bp.
A number of years earlier, Szabó et al. [129] had observed similar products with IS30. Additionally, when the IS30 transposase gene was ablated, the deletion frequency was not only reduced by a factor of 103 but the accompanying deletions were more complex, including large deletions or unidentified plasmid rearrangements.
The detailed observations obtained for ISApl1 led to the model shown in Figure IS3.25 A, which is illustrated with a specific example (iii in Fig. IS3.23) (Fig.IS3.25B).
In this model (Fig.IS3.25) it is envisaged that a short complementary sequence occurring outside the IS involved (Fig.IS3.25 i) and that the DNA strand generated by transposition-associated replication (Fig.IS3.25 ii) switches to the complementary sequence (Fig.IS3.25 iii). This proposed structure is equivalent to a Holliday junction (see for example [130] ) which could be resolved by RuvC (see for example [131]) (Fig.IS3.25 iv).
This reinforces the idea that the protein might intervene during the replicative step of copy-out-paste-in transposition perhaps by interfering with the normal transposition process.
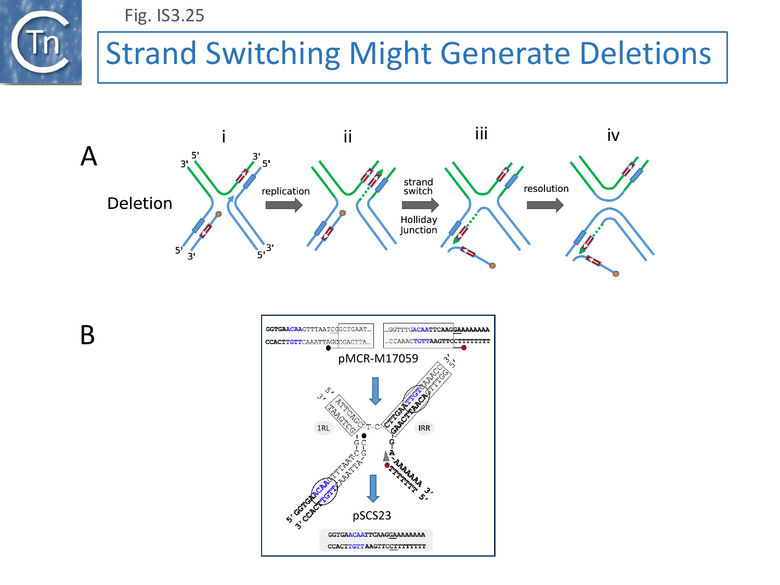
The Primer Invasion model also explains high levels of precise excision.
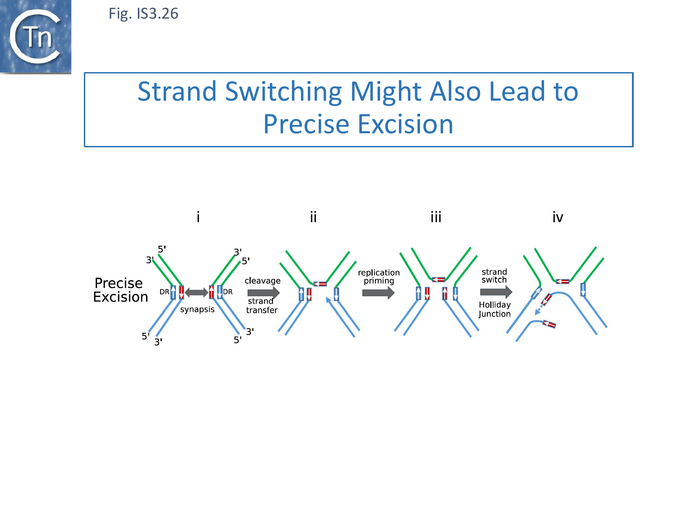
More generally, although the examples of imprecise IS excision led to this model, it also conveniently explains the high level of precise excision observed for IS629 and other IS of the IS3 family [115] (Fig. IS3.26). These generate small 3-4 bp direct flanking target repeats (DR)(Fig. IS3.26) on insertion )(Fig. IS3.22). Transposase-mediated synapsis of the IS ends (Fig. IS3.26 i), cleavage 3-4 nucleotides distal to the opposite IS end (Fig. IS3.26 ii)[85] and single strand bridge formation [85] by strand transfer, includes one strand of the DR, leaving the complementary single-stranded. Priming from the resulting 3’ OH (Fig. IS3.26 iii) would regenerate the second DR strand. Primer invasion (Fig. IS3.26 iv) then permits extension and formation of the Holiday junction. It might be expected that, since the complementary sequences are in proximity, locating microhomologies would be more efficient and therefore precise excision using this type of mechanism would be more frequent than imprecise excision.
This reinforces the idea that the IEE protein might intervene during the replicative step of copy-out-paste-in transposition perhaps by interfering with the normal transposition process.
In summary, the IEE protein plays an important role in excision of members of a number of IS families which all have in common the production of IS circles as transposition intermediates and probably all use the copy-out-paste-in transposition pathway. Excision is not only dependent on IEE but also requires an active transposase, indicating that it is associated with the transposition process itself. Not only do the biochemical properties of IEE include the ability to prime DNA synthesis and overcome potential obstacles due various lesions in the template DNA, they also include the capacity to recognize microhomologies. This suggests to us that IEE acts at the replication (copy-out) step in the transposition pathway, subsequent to initial 3’cleavage of an IS end and its transfer to the other. Specifically, based on sequence data obtained from the loss of the IS30 family member, ISApl1, which flanks the mcr-1 gene in the compound transposon Tn6330, we suggest that it allows a strand switch (primer invasion) to suitable microhomologies in the neighboring donor DNA creating a Holliday junction and short circuiting the replication/transposition reaction. The IS could then be removed by Holliday junction resolution, a possibility that can be addressed experimentally. This would also explain the high levels of precise excision which, by definition leave no scars, since the DR would provide suitable microhomologies to facilitate primer invasion precisely at the IS ends, a prediction which could be tested directly by changing the DR sequence at one end of the IS and examining its effect on precise excision frequency in the presence of IEE.
Importantly, the activity of IEE in removing flanking IS serves to immobilize genes carried by compound transposons, explaining the presence of certain of these genes without associated IS copies in plasmids and chromosomes. It will be important to address this question experimentally using entire compound transposons such as Tn6330 as well as others with flanking copy-out-paste-in IS and to compare the effects with compound transposons with different transposition pathways such as cut-and-paste.
IEE and the Diagnostics Lab
The IS600 excision behavior has been observed in a group of clinically relevant Shiga toxin producing E. coli (E. coli (STEC) O121:H19). It was noted that some, but not all of a number of clinical STEC O121:H19 isolates exhibit a phenotype called delayed lactose utilization (DLU), where cultures remain lac- after 24 h of growth but become lac+ after 48h of growth [132][133].
Excision of IS600 which was inserted into lacZ gene was thought to be responsible and the phenomenon was observed to require the presence of lactose in the medium. Moreover, the resulting lac+ cultures, the IS600 copy had excised. The phenomenon was also shown to require a functional iee gene since its inactivation by an IS1203 copy in a natural isolate prevented DLU. DLU is therefore simply the result of a selection for lac+ individuals in a population with high levels of IS600 excision facilitated by iee.
Acknowledgements
We would like to thank Miguel de Vega (Centro de Biología Molecular Severo Ochoa - Madrid, Spain), Tetsuya Hayashi (Department of Bacteriology, Kyushu University, Fukuoka, Japan) and Bernard Hallet (Universite de Louvain, Louvain la Neuve, Belgium) for reading the Excision section of this chapter and for critical comments.
Bibliography
- ↑ Fiandt M, Szybalski W, Malamy MH . Polar mutations in lac, gal and phage lambda consist of a few IS-DNA sequences inserted with either orientation. - Mol Gen Genet: 1972, 119(3);223-31 [PubMed:4567156] [DOI]
- ↑ Hu S, Ohtsubo E, Davidson N . Electron microscopic heteroduplex studies of sequence relations among plasmids of Escherichia coli: structure of F13 and related F-primes. - J Bacteriol: 1975 May, 122(2);749-63 [PubMed:1092667] [DOI]
- ↑ Hu S, Otsubo E, Davidson N, Saedler H . Electron microscope heteroduplex studies of sequence relations among bacterial plasmids: identification and mapping of the insertion sequences IS1 and IS2 in F and R plasmids. - J Bacteriol: 1975 May, 122(2);764-75 [PubMed:1092668] [DOI]
- ↑ Ptashne K, Cohen SN . Occurrence of insertion sequence (IS) regions on plasmid deoxyribonucleic acid as direct and inverted nucleotide sequence duplications. - J Bacteriol: 1975 May, 122(2);776-81 [PubMed:1092669] [DOI]
- ↑ Ross DG, Grisafi P, Kleckner N, Botstein D . The ends of Tn10 are not IS3. - J Bacteriol: 1979 Sep, 139(3);1097-101 [PubMed:383689] [DOI]
- ↑ Ishiguro N, Sato G, Sasakawa C, Danbara H, Yoshikawa M . Identification of citrate utilization transposon Tn3411 from a naturally occurring citrate utilization plasmid. - J Bacteriol: 1982 Mar, 149(3);961-8 [PubMed:6277857] [DOI]
- ↑ Ishiguro N, Sato G . Nucleotide sequence of insertion sequence IS3411, which flanks the citrate utilization determinant of transposon Tn3411. - J Bacteriol: 1988 Apr, 170(4);1902-6 [PubMed:2832386] [DOI]
- ↑ Ishiguro N, Sato G . Spontaneous deletion of citrate-utilizing ability promoted by insertion sequences. - J Bacteriol: 1984 Nov, 160(2);642-50 [PubMed:6094480] [DOI]
- ↑ Craig NL, Lambowitz AM, Craigie R, Gellert M, editors. Mobile DNA II. American Society of Microbiology; 2002.
- ↑ 10.0 10.1 Chandler M, Fayet O, Rousseau P, Ton Hoang B, Duval-Valentin G . Copy-out-Paste-in Transposition of IS911: A Major Transposition Pathway. - Microbiol Spectr: 2015 Aug, 3(4); [PubMed:26350305] [DOI]
- ↑ 11.0 11.1 11.2 11.3 Prère MF, Chandler M, Fayet O . Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. - J Bacteriol: 1990 Jul, 172(7);4090-9 [PubMed:2163395] [DOI]
- ↑ Blattner FR, Plunkett G 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y . The complete genome sequence of Escherichia coli K-12. - Science: 1997 Sep 5, 277(5331);1453-62 [PubMed:9278503] [DOI]
- ↑ Haren L, Bétermier M, Polard P, Chandler M . IS911-mediated intramolecular transposition is naturally temperature sensitive. - Mol Microbiol: 1997 Aug, 25(3);531-40 [PubMed:9302015] [DOI]
- ↑ Tobe T, Hayashi T, Han CG, Schoolnik GK, Ohtsubo E, Sasakawa C . Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. - Infect Immun: 1999 Oct, 67(10);5455-62 [PubMed:10496929] [DOI]
- ↑ Swenson DL, Bukanov NO, Berg DE, Welch RA . Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. - Infect Immun: 1996 Sep, 64(9);3736-43 [PubMed:8751923] [DOI]
- ↑ Filée J, Siguier P, Chandler M . Insertion sequence diversity in archaea. - Microbiol Mol Biol Rev: 2007 Mar, 71(1);121-57 [PubMed:17347521] [DOI]
- ↑ Smith KS, Ingram-Smith C . Methanosaeta, the forgotten methanogen? - Trends Microbiol: 2007 Apr, 15(4);150-5 [PubMed:17320399] [DOI]
- ↑ 18.0 18.1 Bhugra B, Dybvig K . Identification and characterization of IS1138, a transposable element from Mycoplasma pulmonis that belongs to the IS3 family. - Mol Microbiol: 1993 Feb, 7(4);577-84 [PubMed:8096321] [DOI]
- ↑ 19.0 19.1 19.2 19.3 19.4 Vögele K, Schwartz E, Welz C, Schiltz E, Rak B . High-level ribosomal frameshifting directs the synthesis of IS150 gene products. - Nucleic Acids Res: 1991 Aug 25, 19(16);4377-85 [PubMed:1653413] [DOI]
- ↑ 20.0 20.1 20.2 20.3 20.4 20.5 20.6 Sekine Y, Eisaki N, Ohtsubo E . Translational control in production of transposase and in transposition of insertion sequence IS3. - J Mol Biol: 1994 Feb 4, 235(5);1406-20 [PubMed:8107082] [DOI]
- ↑ 21.0 21.1 21.2 21.3 21.4 21.5 21.6 21.7 21.8 Polard P, Prère MF, Chandler M, Fayet O . Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. - J Mol Biol: 1991 Dec 5, 222(3);465-77 [PubMed:1660923] [DOI]
- ↑ 22.0 22.1 22.2 22.3 22.4 Lewis LA, Grindley ND . Two abundant intramolecular transposition products, resulting from reactions initiated at a single end, suggest that IS2 transposes by an unconventional pathway. - Mol Microbiol: 1997 Aug, 25(3);517-29 [PubMed:9302014] [DOI]
- ↑ Chandler M, Fayet O . Translational frameshifting in the control of transposition in bacteria. - Mol Microbiol: 1993 Feb, 7(4);497-503 [PubMed:8384687] [DOI]
- ↑ 24.0 24.1 Sharma V, Firth AE, Antonov I, Fayet O, Atkins JF, Borodovsky M, Baranov PV . A pilot study of bacterial genes with disrupted ORFs reveals a surprising profusion of protein sequence recoding mediated by ribosomal frameshifting and transcriptional realignment. - Mol Biol Evol: 2011 Nov, 28(11);3195-211 [PubMed:21673094] [DOI]
- ↑ 25.0 25.1 Sharma V, Prère MF, Canal I, Firth AE, Atkins JF, Baranov PV, Fayet O . Analysis of tetra- and hepta-nucleotides motifs promoting -1 ribosomal frameshifting in Escherichia coli. - Nucleic Acids Res: 2014 Jun, 42(11);7210-25 [PubMed:24875478] [DOI]
- ↑ 26.0 26.1 26.2 26.3 Hu ST, Hwang JH, Lee LC, Lee CH, Li PL, Hsieh YC . Functional analysis of the 14 kDa protein of insertion sequence 2. - J Mol Biol: 1994 Feb 18, 236(2);503-13 [PubMed:8107136] [DOI]
- ↑ Reimmann C, Moore R, Little S, Savioz A, Willetts NS, Haas D . Genetic structure, function and regulation of the transposable element IS21. - Mol Gen Genet: 1989 Feb, 215(3);416-24 [PubMed:2540414] [DOI]
- ↑ Dalrymple B . Novel rearrangements of IS30 carrying plasmids leading to the reactivation of gene expression. - Mol Gen Genet: 1987 May, 207(2-3);413-20 [PubMed:3039299] [DOI]
- ↑ Perkins-Balding D, Duval-Valentin G, Glasgow AC . Excision of IS492 requires flanking target sequences and results in circle formation in Pseudoalteromonas atlantica. - J Bacteriol: 1999 Aug, 181(16);4937-48 [PubMed:10438765] [DOI]
- ↑ 30.0 30.1 30.2 30.3 Duval-Valentin G, Normand C, Khemici V, Marty B, Chandler M . Transient promoter formation: a new feedback mechanism for regulation of IS911 transposition. - EMBO J: 2001 Oct 15, 20(20);5802-11 [PubMed:11598022] [DOI]
- ↑ 31.0 31.1 Spielmann-Ryser J, Moser M, Kast P, Weber H . Factors determining the frequency of plasmid cointegrate formation mediated by insertion sequence IS3 from Escherichia coli. - Mol Gen Genet: 1991 May, 226(3);441-8 [PubMed:1645443] [DOI]
- ↑ Sengstag C, Arber W . A cloned DNA fragment from bacteriophage P1 enhances IS2 insertion. - Mol Gen Genet: 1987 Feb, 206(2);344-51 [PubMed:3035338] [DOI]
- ↑ 33.0 33.1 Polard P, Seroude L, Fayet O, Prère MF, Chandler M . One-ended insertion of IS911. - J Bacteriol: 1994 Feb, 176(4);1192-6 [PubMed:8106332] [DOI]
- ↑ Welz C. Functionelle analyse des Bakteriellen Insertionelements IS150. PhD thesis: Fakultät für Biologie der Albert-Ludwigs-Univesität Freiburg; 1993.
- ↑ Bachellier S, Clément JM, Hofnung M, Gilson E . Bacterial interspersed mosaic elements (BIMEs) are a major source of sequence polymorphism in Escherichia coli intergenic regions including specific associations with a new insertion sequence. - Genetics: 1997 Mar, 145(3);551-62 [PubMed:9055066] [DOI]
- ↑ Ferat JL, Le Gouar M, Michel F . Multiple group II self-splicing introns in mobile DNA from Escherichia coli. - C R Acad Sci III: 1994 Feb, 317(2);141-8 [PubMed:7994604]
- ↑ 37.0 37.1 Mahillon J, Chandler M . Insertion sequences. - Microbiol Mol Biol Rev: 1998 Sep, 62(3);725-74 [PubMed:9729608] [DOI]
- ↑ 38.0 38.1 38.2 38.3 38.4 38.5 38.6 38.7 Haren L, Normand C, Polard P, Alazard R, Chandler M . IS911 transposition is regulated by protein-protein interactions via a leucine zipper motif. - J Mol Biol: 2000 Feb 25, 296(3);757-68 [PubMed:10677279] [DOI]
- ↑ Osawa S, Jukes TH, Watanabe K, Muto A . Recent evidence for evolution of the genetic code. - Microbiol Rev: 1992 Mar, 56(1);229-64 [PubMed:1579111] [DOI]
- ↑ 40.0 40.1 Zheng J, McIntosh MA . Characterization of IS1221 from Mycoplasma hyorhinis: expression of its putative transposase in Escherichia coli incorporates a ribosomal frameshift mechanism. - Mol Microbiol: 1995 May, 16(4);669-85 [PubMed:7476162] [DOI]
- ↑ Petrova M, Shcherbatova N, Gorlenko Z, Mindlin S . A new subgroup of the IS3 family and properties of its representative member ISPpy1. - Microbiology (Reading): 2013 Sep, 159(Pt 9);1900-1910 [PubMed:23832000] [DOI]
- ↑ 42.0 42.1 Doak TG, Doerder FP, Jahn CL, Herrick G . A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common "D35E" motif. - Proc Natl Acad Sci U S A: 1994 Feb 1, 91(3);942-6 [PubMed:8302872] [DOI]
- ↑ 43.0 43.1 43.2 Fayet O, Ramond P, Polard P, Prère MF, Chandler M . Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? - Mol Microbiol: 1990 Oct, 4(10);1771-7 [PubMed:1963920] [DOI]
- ↑ 44.0 44.1 Katzman M, Mack JP, Skalka AM, Leis J . A covalent complex between retroviral integrase and nicked substrate DNA. - Proc Natl Acad Sci U S A: 1991 Jun 1, 88(11);4695-9 [PubMed:1647013] [DOI]
- ↑ 45.0 45.1 Khan E, Mack JP, Katz RA, Kulkosky J, Skalka AM . Retroviral integrase domains: DNA binding and the recognition of LTR sequences. - Nucleic Acids Res: 1991 Feb 25, 19(4);851-60 [PubMed:1850126] [DOI]
- ↑ 46.0 46.1 Rezsöhazy R, Hallet B, Delcour J, Mahillon J . The IS4 family of insertion sequences: evidence for a conserved transposase motif. - Mol Microbiol: 1993 Sep, 9(6);1283-95 [PubMed:7934941] [DOI]
- ↑ Fu R, Voordouw G . ISD1, an insertion element from the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough: structure, transposition, and distribution. - Appl Environ Microbiol: 1998 Jan, 64(1);53-61 [PubMed:9435062] [DOI]
- ↑ 48.0 48.1 Schwartz E, Kröger M, Rak B . IS150: distribution, nucleotide sequence and phylogenetic relationships of a new E. coli insertion element. - Nucleic Acids Res: 1988 Jul 25, 16(14B);6789-802 [PubMed:2841644] [DOI]
- ↑ 49.0 49.1 49.2 Rousseau P, Gueguen E, Duval-Valentin G, Chandler M . The helix-turn-helix motif of bacterial insertion sequence IS911 transposase is required for DNA binding. - Nucleic Acids Res: 2004, 32(4);1335-44 [PubMed:14981152] [DOI]
- ↑ 50.0 50.1 50.2 50.3 50.4 50.5 50.6 50.7 Haren L, Polard P, Ton-Hoang B, Chandler M . Multiple oligomerisation domains in the IS911 transposase: a leucine zipper motif is essential for activity. - J Mol Biol: 1998, 283(1);29-41 [PubMed:9761671] [DOI]
- ↑ Haren L, Ton-Hoang B, Chandler M . Integrating DNA: transposases and retroviral integrases. - Annu Rev Microbiol: 1999, 53;245-81 [PubMed:10547692] [DOI]
- ↑ 52.0 52.1 52.2 Rettberg CC, Prère MF, Gesteland RF, Atkins JF, Fayet O . A three-way junction and constituent stem-loops as the stimulator for programmed -1 frameshifting in bacterial insertion sequence IS911. - J Mol Biol: 1999 Mar 12, 286(5);1365-78 [PubMed:10064703] [DOI]
- ↑ 53.0 53.1 Prère MF, Canal I, Wills NM, Atkins JF, Fayet O . The interplay of mRNA stimulatory signals required for AUU-mediated initiation and programmed -1 ribosomal frameshifting in decoding of transposable element IS911. - J Bacteriol: 2011 Jun, 193(11);2735-44 [PubMed:21478364] [DOI]
- ↑ 54.0 54.1 54.2 54.3 Mazauric MH, Licznar P, Prère MF, Canal I, Fayet O . Apical loop-internal loop RNA pseudoknots: a new type of stimulator of -1 translational frameshifting in bacteria. - J Biol Chem: 2008 Jul 18, 283(29);20421-32 [PubMed:18474594] [DOI]
- ↑ Chen CC, Hu ST . Two frameshift products involved in the transposition of bacterial insertion sequence IS629. - J Biol Chem: 2006 Aug 4, 281(31);21617-21628 [PubMed:16731525] [DOI]
- ↑ 56.0 56.1 Hu ST, Lee LC, Lei GS . Detection of an IS2-encoded 46-kilodalton protein capable of binding terminal repeats of IS2. - J Bacteriol: 1996 Oct, 178(19);5652-9 [PubMed:8824609] [DOI]
- ↑ Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM . Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. - Mol Cell Biol: 1992 May, 12(5);2331-8 [PubMed:1314954] [DOI]
- ↑ Brierley I . Ribosomal frameshifting viral RNAs. - J Gen Virol: 1995 Aug, 76 ( Pt 8);1885-92 [PubMed:7636469] [DOI]
- ↑ 59.0 59.1 Tsuchihashi Z, Brown PO . Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(Lys) and an AAG lysine codon. - Genes Dev: 1992 Mar, 6(3);511-9 [PubMed:1547945] [DOI]
- ↑ Yokoyama S, Watanabe T, Murao K, Ishikura H, Yamaizumi Z, Nishimura S, Miyazawa T . Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. - Proc Natl Acad Sci U S A: 1985 Aug, 82(15);4905-9 [PubMed:3860833] [DOI]
- ↑ Sundaram M, Durant PC, Davis DR . Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. - Biochemistry: 2000 Oct 17, 39(41);12575-84 [PubMed:11027137] [DOI]
- ↑ Licznar P, Mejlhede N, Prère MF, Wills N, Gesteland RF, Atkins JF, Fayet O . Programmed translational -1 frameshifting on hexanucleotide motifs and the wobble properties of tRNAs. - EMBO J: 2003 Sep 15, 22(18);4770-8 [PubMed:12970189] [DOI]
- ↑ Giedroc DP, Cornish PV . Frameshifting RNA pseudoknots: structure and mechanism. - Virus Res: 2009 Feb, 139(2);193-208 [PubMed:18621088] [DOI]
- ↑ 64.0 64.1 Licznar P, Bertrand C, Canal I, Prère MF, Fayet O . Genetic variability of the frameshift region in IS911 transposable elements from Escherichia coli clinical isolates. - FEMS Microbiol Lett: 2003 Jan 28, 218(2);231-7 [PubMed:12586397] [DOI]
- ↑ 65.0 65.1 65.2 65.3 Duval-Valentin G, Chandler M . Cotranslational control of DNA transposition: a window of opportunity. - Mol Cell: 2011 Dec 23, 44(6);989-96 [PubMed:22195971] [DOI]
- ↑ Utsumi R, Ikeda M, Horie T, Yamamoto M, Ichihara A, Taniguchi Y, Hashimoto R, Tanabe H, Obata K, Noda M . Isolation and characterization of the IS3-like element from Thermus aquaticus. - Biosci Biotechnol Biochem: 1995 Sep, 59(9);1707-11 [PubMed:8520113] [DOI]
- ↑ Boursaux-Eude C, Saint Girons I, Zuerner R . IS1500, an IS3-like element from Leptospira interrogans. - Microbiology (Reading): 1995 Sep, 141 ( Pt 9);2165-73 [PubMed:7496528] [DOI]
- ↑ Lei GS, Hu ST . Functional domains of the InsA protein of IS2. - J Bacteriol: 1997 Oct, 179(20);6238-43 [PubMed:9335268] [DOI]
- ↑ Haren L, Polard P, Ton-Hoang B, Chandler M . Multiple oligomerisation domains in the IS911 transposase: a leucine zipper motif is essential for activity. - J Mol Biol: 1998, 283(1);29-41 [PubMed:9761671] [DOI]
- ↑ Shieh YW, Minguez P, Bork P, Auburger JJ, Guilbride DL, Kramer G, Bukau B . Operon structure and cotranslational subunit association direct protein assembly in bacteria. - Science: 2015 Nov 6, 350(6261);678-80 [PubMed:26405228] [DOI]
- ↑ Montaño SP, Rice PA . Moving DNA around: DNA transposition and retroviral integration. - Curr Opin Struct Biol: 2011 Jun, 21(3);370-8 [PubMed:21439812] [DOI]
- ↑ Dyda F, Chandler M, Hickman AB . The emerging diversity of transpososome architectures. - Q Rev Biophys: 2012 Nov, 45(4);493-521 [PubMed:23217365] [DOI]
- ↑ Gueguen E, Rousseau P, Duval-Valentin G, Chandler M . The transpososome: control of transposition at the level of catalysis. - Trends Microbiol: 2005 Nov, 13(11);543-9 [PubMed:16181782] [DOI]
- ↑ Normand C, Duval-Valentin G, Haren L, Chandler M . The terminal inverted repeats of IS911: requirements for synaptic complex assembly and activity. - J Mol Biol: 2001 May 18, 308(5);853-71 [PubMed:11352577] [DOI]
- ↑ 75.0 75.1 Rousseau P, Tardin C, Tolou N, Salomé L, Chandler M . A model for the molecular organisation of the IS911 transpososome. - Mob DNA: 2010 Jun 16, 1(1);16 [PubMed:20553579] [DOI]
- ↑ Pouget N, Dennis C, Turlan C, Grigoriev M, Chandler M, Salomé L . Single-particle tracking for DNA tether length monitoring. - Nucleic Acids Res: 2004 May 20, 32(9);e73 [PubMed:15155821] [DOI]
- ↑ Pouget N, Turlan C, Destainville N, Salomé L, Chandler M . IS911 transpososome assembly as analysed by tethered particle motion. - Nucleic Acids Res: 2006, 34(16);4313-23 [PubMed:16923775] [DOI]
- ↑ 78.0 78.1 78.2 Rousseau P, Loot C, Guynet C, Ah-Seng Y, Ton-Hoang B, Chandler M . Control of IS911 target selection: how OrfA may ensure IS dispersion. - Mol Microbiol: 2007 Mar, 63(6);1701-9 [PubMed:17367389] [DOI]
- ↑ Rousseau P, Loot C, Turlan C, Nolivos S, Chandler M . Bias between the left and right inverted repeats during IS911 targeted insertion. - J Bacteriol: 2008 Sep, 190(18);6111-8 [PubMed:18586933] [DOI]
- ↑ 80.0 80.1 80.2 Ton-Hoang B, Polard P, Chandler M . Efficient transposition of IS911 circles in vitro. - EMBO J: 1998 Feb 16, 17(4);1169-81 [PubMed:9463394] [DOI]
- ↑ 81.0 81.1 81.2 Turlan C, Chandler M . IS1-mediated intramolecular rearrangements: formation of excised transposon circles and replicative deletions. - EMBO J: 1995 Nov 1, 14(21);5410-21 [PubMed:7489730] [DOI]
- ↑ Ohtsubo E, Minematsu H, Tsuchida K, Ohtsubo H, Sekine Y . Intermediate molecules generated by transposase in the pathways of transposition of bacterial insertion element IS3. - Adv Biophys: 2004, 38;125-39 [PubMed:15493331]
- ↑ 83.0 83.1 Haas M, Rak B . Escherichia coli insertion sequence IS150: transposition via circular and linear intermediates. - J Bacteriol: 2002 Nov, 184(21);5833-41 [PubMed:12374815] [DOI]
- ↑ Hickman AB, Dyda F . Mechanisms of DNA Transposition. - Microbiol Spectr: 2015 Apr, 3(2);MDNA3-0034-2014 [PubMed:26104718] [DOI]
- ↑ 85.0 85.1 85.2 85.3 85.4 Polard P, Chandler M . An in vivo transposase-catalyzed single-stranded DNA circularization reaction. - Genes Dev: 1995 Nov 15, 9(22);2846-58 [PubMed:7590258] [DOI]
- ↑ 86.0 86.1 86.2 Sekine Y, Aihara K, Ohtsubo E . Linearization and transposition of circular molecules of insertion sequence IS3. - J Mol Biol: 1999 Nov 19, 294(1);21-34 [PubMed:10556026] [DOI]
- ↑ Sekine Y, Izumi K, Mizuno T, Ohtsubo E . Inhibition of transpositional recombination by OrfA and OrfB proteins encoded by insertion sequence IS3. - Genes Cells: 1997 Sep, 2(9);547-57 [PubMed:9413996] [DOI]
- ↑ Polard P, Prère MF, Fayet O, Chandler M . Transposase-induced excision and circularization of the bacterial insertion sequence IS911. - EMBO J: 1992 Dec, 11(13);5079-90 [PubMed:1334464] [DOI]
- ↑ 89.0 89.1 89.2 Duval-Valentin G, Marty-Cointin B, Chandler M . Requirement of IS911 replication before integration defines a new bacterial transposition pathway. - EMBO J: 2004 Oct 1, 23(19);3897-906 [PubMed:15359283] [DOI]
- ↑ Bierne H, Ehrlich SD, Michel B . Flanking sequences affect replication arrest at the Escherichia coli terminator TerB in vivo. - J Bacteriol: 1994 Jul, 176(13);4165-7 [PubMed:8021197] [DOI]
- ↑ Hill TM, Marians KJ . Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. - Proc Natl Acad Sci U S A: 1990 Apr, 87(7);2481-5 [PubMed:2181438] [DOI]
- ↑ Kuempel PL, Pelletier AJ, Hill TM . Tus and the terminators: the arrest of replication in prokaryotes. - Cell: 1989 Nov 17, 59(4);581-3 [PubMed:2510933] [DOI]
- ↑ Neylon C, Kralicek AV, Hill TM, Dixon NE . Replication termination in Escherichia coli: structure and antihelicase activity of the Tus-Ter complex. - Microbiol Mol Biol Rev: 2005 Sep, 69(3);501-26 [PubMed:16148308] [DOI]
- ↑ Harshey RM . Transposable Phage Mu. - Microbiol Spectr: 2014 Oct, 2(5); [PubMed:26104374] [DOI]
- ↑ Burton BM, Baker TA . Mu transpososome architecture ensures that unfolding by ClpX or proteolysis by ClpXP remodels but does not destroy the complex. - Chem Biol: 2003 May, 10(5);463-72 [PubMed:12770828] [DOI]
- ↑ Nakai H, Doseeva V, Jones JM . Handoff from recombinase to replisome: insights from transposition. - Proc Natl Acad Sci U S A: 2001 Jul 17, 98(15);8247-54 [PubMed:11459960] [DOI]
- ↑ 97.0 97.1 97.2 Turlan C, Loot C, Chandler M . IS911 partial transposition products and their processing by the Escherichia coli RecG helicase. - Mol Microbiol: 2004 Aug, 53(4);1021-33 [PubMed:15306008] [DOI]
- ↑ 98.0 98.1 98.2 Ton-Hoang B, Bétermier M, Polard P, Chandler M . Assembly of a strong promoter following IS911 circularization and the role of circles in transposition. - EMBO J: 1997 Jun 2, 16(11);3357-71 [PubMed:9214651] [DOI]
- ↑ Lewis LA, Cylin E, Lee HK, Saby R, Wong W, Grindley ND . The left end of IS2: a compromise between transpositional activity and an essential promoter function that regulates the transposition pathway. - J Bacteriol: 2004 Feb, 186(3);858-65 [PubMed:14729714] [DOI]
- ↑ Ton-Hoang B, Polard P, Haren L, Turlan C, Chandler M . IS911 transposon circles give rise to linear forms that can undergo integration in vitro. - Mol Microbiol: 1999 May, 32(3);617-27 [PubMed:10320583] [DOI]
- ↑ 101.0 101.1 Loot C, Turlan C, Rousseau P, Ton-Hoang B, Chandler M . A target specificity switch in IS911 transposition: the role of the OrfA protein. - EMBO J: 2002 Aug 1, 21(15);4172-82 [PubMed:12145217] [DOI]
- ↑ Loot C, Turlan C, Chandler M . Host processing of branched DNA intermediates is involved in targeted transposition of IS911. - Mol Microbiol: 2004 Jan, 51(2);385-93 [PubMed:14756780] [DOI]
- ↑ 103.0 103.1 Sekine Y, Eisaki N, Ohtsubo E . Identification and characterization of the linear IS3 molecules generated by staggered breaks. - J Biol Chem: 1996 Jan 5, 271(1);197-202 [PubMed:8550559] [DOI]
- ↑ Ghosal D, Sommer H, Saedler H . Nucleotide sequence of the transposable DNA-element IS2. - Nucleic Acids Res: 1979 Mar, 6(3);1111-22 [PubMed:375194] [DOI]
- ↑ Szeverényi I, Bodoky T, Olasz F . Isolation, characterization and transposition of an (IS2)2 intermediate. - Mol Gen Genet: 1996 Jun 12, 251(3);281-9 [PubMed:8676870] [DOI]
- ↑ Kosek D, Hickman AB, Ghirlando R, He S, Dyda F . Structures of ISCth4 transpososomes reveal the role of asymmetry in copy-out/paste-in DNA transposition. - EMBO J: 2021 Jan 4, 40(1);e105666 [PubMed:33006208] [DOI]
- ↑ Ton-Hoang B, Polard P, Chandler M . Efficient transposition of IS911 circles in vitro. - EMBO J: 1998 Feb 16, 17(4);1169-81 [PubMed:9463394] [DOI]
- ↑ Chandler M, Ross K, Varani AM . The insertion sequence excision enhancer: A PrimPol-based primer invasion system for immobilizing transposon-transmitted antibiotic resistance genes. - Mol Microbiol: 2023 Aug 13; [PubMed:37574851] [DOI]
- ↑ Kusumoto M, Nishiya Y, Kawamura Y . Reactivation of insertionally inactivated Shiga toxin 2 genes of Escherichia coli O157:H7 caused by nonreplicative transposition of the insertion sequence. - Appl Environ Microbiol: 2000 Mar, 66(3);1133-8 [PubMed:10698782] [DOI]
- ↑ Kusumoto M, Suzuki R, Nishiya Y, Okitsu T, Oka M . Host-dependent activation of IS1203v excision in Shiga toxin-producing Escherichia coli. - J Biosci Bioeng: 2004, 97(6);406-11 [PubMed:16233651] [DOI]
- ↑ Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H . Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. - DNA Res: 2001 Feb 28, 8(1);11-22 [PubMed:11258796] [DOI]
- ↑ Makino K, Ishii K, Yasunaga T, Hattori M, Yokoyama K, Yutsudo CH, Kubota Y, Yamaichi Y, Iida T, Yamamoto K, Honda T, Han CG, Ohtsubo E, Kasamatsu M, Hayashi T, Kuhara S, Shinagawa H . Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. - DNA Res: 1998 Feb 28, 5(1);1-9 [PubMed:9628576] [DOI]
- ↑ Kusumoto M, Suzuki R, Nishiya Y, Okitsu T, Oka M . Host-dependent activation of IS1203v excision in Shiga toxin-producing Escherichia coli. - J Biosci Bioeng: 2004, 97(6);406-11 [PubMed:16233651] [DOI]
- ↑ 114.0 114.1 114.2 Kusumoto M, Fukamizu D, Ogura Y, Yoshida E, Yamamoto F, Iwata T, Ooka T, Akiba M, Hayashi T . Lineage-specific distribution of insertion sequence excision enhancer in enterotoxigenic Escherichia coli isolated from swine. - Appl Environ Microbiol: 2014 Feb, 80(4);1394-402 [PubMed:24334665] [DOI]
- ↑ 115.00 115.01 115.02 115.03 115.04 115.05 115.06 115.07 115.08 115.09 115.10 115.11 115.12 115.13 115.14 115.15 Kusumoto M, Ooka T, Nishiya Y, Ogura Y, Saito T, Sekine Y, Iwata T, Akiba M, Hayashi T . Insertion sequence-excision enhancer removes transposable elements from bacterial genomes and induces various genomic deletions. - Nat Commun: 2011 Jan 11, 2;152 [PubMed:21224843] [DOI]
- ↑ 116.0 116.1 116.2 116.3 116.4 116.5 116.6 116.7 116.8 116.9 Calvo PA, Mateo-Cáceres V, Díaz-Arco S, Redrejo-Rodríguez M, de Vega M . The enterohemorrhagic Escherichia coli insertion sequence-excision enhancer protein is a DNA polymerase with microhomology-mediated end-joining activity. - Nucleic Acids Res: 2023 Jan 30; [PubMed:36715333] [DOI]
- ↑ Lundblad V, Kleckner N . Mutants of Escherichia coli K12 which affect excision of transposon Tn10. - Basic Life Sci: 1982, 20;245-58 [PubMed:6287993] [DOI]
- ↑ Lundblad V, Taylor AF, Smith GR, Kleckner N . Unusual alleles of recB and recC stimulate excision of inverted repeat transposons Tn10 and Tn5. - Proc Natl Acad Sci U S A: 1984 Feb, 81(3);824-8 [PubMed:6322169] [DOI]
- ↑
Lundblad V, Kleckner N . Mismatch repair mutations of Escherichia coli K12 enhance transposon excision. - Genetics: 1985 Jan, 109(1);3-19 [PubMed:2981756]
[DOI]
- ↑ Benjamin HW, Kleckner N . Excision of Tn10 from the donor site during transposition occurs by flush double-strand cleavages at the transposon termini. - Proc Natl Acad Sci U S A: 1992 May 15, 89(10);4648-52 [PubMed:1316613] [DOI]
- ↑ Szabó M, Kiss J, Nagy Z, Chandler M, Olasz F . Sub-terminal sequences modulating IS30 transposition in vivo and in vitro. - J Mol Biol: 2008 Jan 11, 375(2);337-52 [PubMed:18022196] [DOI]
- ↑ Ross DG, Swan J, Kleckner N . Nearly precise excision: a new type of DNA alteration associated with the translocatable element Tn10. - Cell: 1979 Apr, 16(4);733-8 [PubMed:455447] [DOI]
- ↑ Foster TJ, Lundblad V, Hanley-Way S, Halling SM, Kleckner N . Three Tn10-associated excision events: relationship to transposition and role of direct and inverted repeats. - Cell: 1981 Jan, 23(1);215-27 [PubMed:6260376] [DOI]
- ↑ 124.0 124.1 124.2 124.3 Snesrud E, McGann P, Chandler M . The Birth and Demise of the ISApl1-mcr-1-ISApl1 Composite Transposon: the Vehicle for Transferable Colistin Resistance. - mBio: 2018 Feb 13, 9(1); [PubMed:29440577] [DOI]
- ↑ 125.0 125.1 Snesrud E, He S, Chandler M, Dekker JP, Hickman AB, McGann P, Dyda F . A Model for Transposition of the Colistin Resistance Gene mcr-1 by ISApl1. - Antimicrob Agents Chemother: 2016 Nov, 60(11);6973-6976 [PubMed:27620479] [DOI]
- ↑ Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J . Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. - Lancet Infect Dis: 2016 Feb, 16(2);161-8 [PubMed:26603172] [DOI]
- ↑ Li R, Xie M, Zhang J, Yang Z, Liu L, Liu X, Zheng Z, Chan EW, Chen S . Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. - J Antimicrob Chemother: 2017 Feb, 72(2);393-401 [PubMed:28073961] [DOI]
- ↑ Poirel L, Kieffer N, Nordmann P . In Vitro Study of ISApl1-Mediated Mobilization of the Colistin Resistance Gene mcr-1. - Antimicrob Agents Chemother: 2017 Jul, 61(7); [PubMed:28416554] [DOI]
- ↑ Szabó M, Kiss J, Kótány G, Olasz F . Importance of illegitimate recombination and transposition in IS30-associated excision events. - Plasmid: 1999 Nov, 42(3);192-209 [PubMed:10545262] [DOI]
- ↑ Lilley DMJ . Holliday junction-resolving enzymes-structures and mechanisms. - FEBS Lett: 2017 Apr, 591(8);1073-1082 [PubMed:27990631] [DOI]
- ↑ Kuzminov A . RuvA, RuvB and RuvC proteins: cleaning-up after recombinational repairs in E. coli. - Bioessays: 1993 May, 15(5);355-8 [PubMed:8393667] [DOI]
- ↑ Nakamura K, Seto K, Isobe J, Taniguchi I, Gotoh Y, Hayashi T . Insertion Sequence (IS)-Excision Enhancer (IEE)-Mediated IS Excision from the lacZ Gene Restores the Lactose Utilization Defect of Shiga Toxin-Producing Escherichia coli O121:H19 Strains and Is Responsible for Their Delayed Lactose Utilization Phenotype. - Appl Environ Microbiol: 2022 Aug 23, 88(16);e0076022 [PubMed:35913153] [DOI]
- ↑ Gill A, McMahon T, Dussault F, Jinneman K, Lindsey R, Martin H, Stoneburg D, Strockbine N, Wetherington J, Feng P . Delayed lactose utilization among Shiga toxin-producing Escherichia coli of serogroup O121. - Food Microbiol: 2022 Apr, 102;103903 [PubMed:34809935] [DOI]