General Information/IS Organization
Contents
General
In addition to being small, insertion sequences are genetically compact (Fig.18.1). They generally encode no functions other than those involved in their mobility, although individual members of several families which include additional genes are now being identified. IS-encoded functions include factors required in cis, in particular recombinationally active DNA sequences that define the ends of the element together with an enzyme, the transposase (Tpase), which recognises and processes these ends. The Tpase is generally encoded by a single, or perhaps two, open reading frames and consumes nearly the entire length of the element.
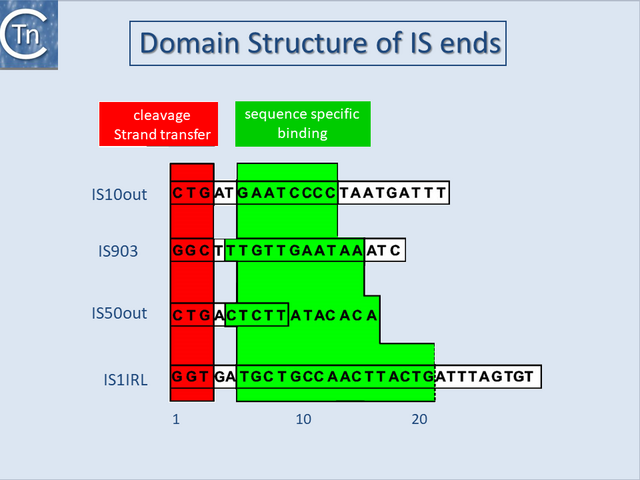
Terminal inverted repeats
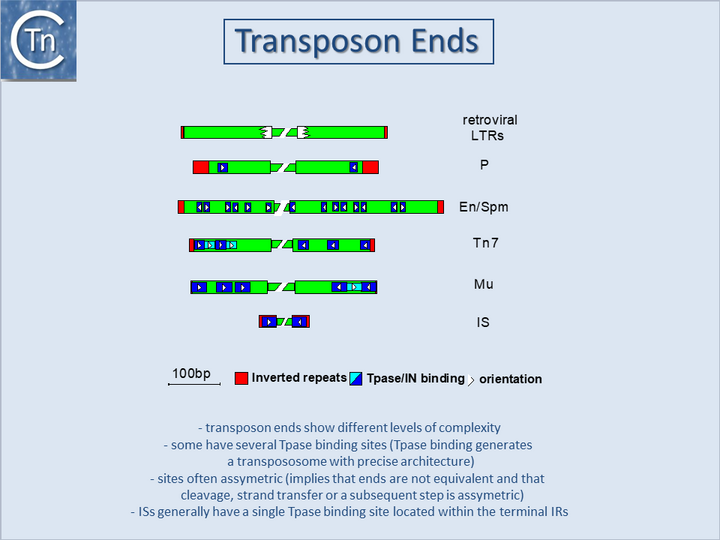
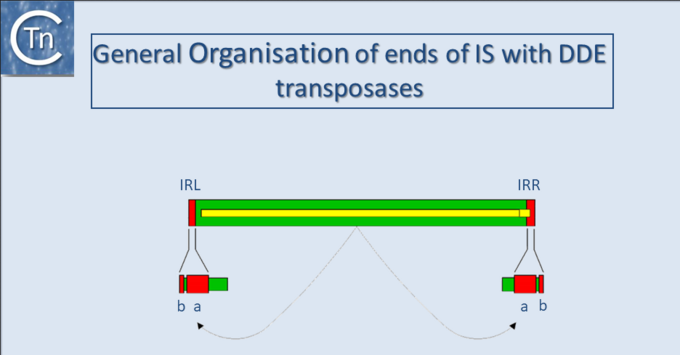
With several notable exceptions (the IS91, IS110 and IS200/605 families; TABLE Characteristics of IS families) the majority of ISs exhibit short terminal IR of between 10 and 40 bp. In those cases examined experimentally, the IRs can be divided into two functional domains (Fig.11.1). Domain “b” includes the two or three terminal base pairs (Fig.18.2), and is involved in the cleavages and strand transfer reactions leading to transposition of the element. Domain “a” is positioned within the IR and is involved in Tpase binding[1][2][3][4][5][6][7].
A similar organization has also been proposed for the transposon Tn3[8] and for the related γδ transposon[9]. The simple single terminal Tpase binding sites of ISs are to be contrasted with the multiple and asymmetric protein binding sites observed in the case of bacteriophage Mu[10] and transposons Tn7[11], and probably Tn552[12][13]. Multiple protein binding sites are also a characteristic of the complex En/Spm and Ac elements of maize (see [14][15] (Fig.18.3). It is worth noting that members of the IS21 family also carry multiple repeated sequences at both ends, which may also represent Tpase binding sites[16][17].
By accommodating different binding patterns at each end, such an arrangement can provide a functional distinction between the ends, either in the assembly or in the activity of the synaptic complex. In addition, indigenous IS promoters are often located partially within the IR sequence upstream of the Tpase gene, by convention IRL. This arrangement may provide a mechanism for autoregulation of Tpase synthesis by Tpase binding. Binding sites for host specified proteins are also often found within or close to the terminal IRs and these proteins may play a role in modulating transposition activity or Tpase expression.
Domain structure of transposases
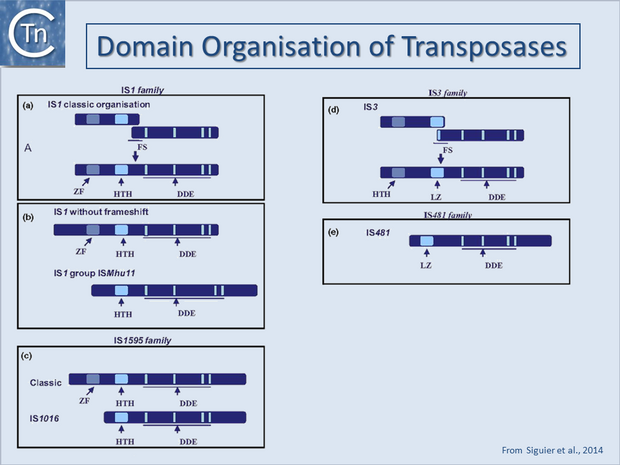
A general pattern for the functional organisation of Tpases appears to be emerging from the increasing number which have been analysed. Many can be divided into topologically distinct structural domains and, although several regions of the protein may contribute to a given function, the isolated domains themselves often exhibit a distinct function. The sequence-specific DNA binding activities of the proteins are generally located in the N-terminal region while the catalytic domain is often localised towards the C-terminal end: IS1[18][19]; IS30[20];Mu, (see [21][22]; Tn3[23][24]; IS50[25]; IS903[26]; IS911[27]; for a review see [28] (Fig.18.4). One functional interpretation of this arrangement for prokaryotic elements is that it may permit interaction of a nascent protein molecule with its target sequences on the IS thus coupling expression and activity. This notion is reinforced by the observation that the presence of the C-terminal region of the IS50, IS10 and IS911 Tpases appears to mask the DNA binding domain and reduce binding activity[29][30] possibly by masking the DNA binding domain. This arrangement might favor activity of the protein in cis, a property shared by several Tpases (see Activity in cis). Similar masking appears to occur with the IS1 (D. Zerbib and M. Chandler, unpublished) and the IS911[31][7] Tpases. In several cases these domains are assembled into a single protein from consecutive orfs by translational frameshifting (Programmed translational frameshifting). In the case of IS911, it has been demonstrated that transposase binding to the IS ends occurs as the protein is translated[32].
One exception to this is the transposase of the IS110 family which encodes a DEDD transposase closely related to the RuvC Holiday resolvase (see [33][34]) and in which the catalytic domain appears to precede the DNA binding domain.
In addition to functional domains for DNA binding and catalysis, many, if not all transposases have the capacity to generate multimeric forms essential for their activity (see [28][35]). This is true of prokaryotic elements such as bacteriophage Mu (see [36]), IS50[37], IS911[31][7][35], IS608 and ISDra2[38][39] (but apparently not IS10[40], and of eukaryotic elements such as the retroviruses (see [41][42][43][44]) whose integrase (IN) (transposase) appears to be a dimer of dimers both with and without DNA bound[43][45][46][47] as does the purified P element transposase[48], the mariner-like element, Mos1[49][50][51] and hermes (which appears to be an octomer)[52][53][54]. With the results of an increasing number of structural studies of these types of enzyme, it will be of great interest to compare the overall similarities of equivalent functional domains as has been recently possible with the catalytic domains of retroviral integrases, Mu transposase and other polynucleotidyl transferases such as the Holiday resolvase, RuvC and RnaseH (see [55][56]). One particular type of structure, a leucine zipper, clearly plays a vital role in multimerization of a number of transposases[31][35][57][58][59][60][61][62].
Finally, several DDE transposases exhibit what may be thought of as an “orphan” domain located between the second D and final E of the DDE motif[63] (Table transposases examined by secondary structure prediction programs). These can be either largely α-helical or β-stranded. It is likely that these insertion domains play subtle roles in the chemical pathways involved in the transposition of their cognate transposable elements. The largely β-stranded IS50 (Tn5) associated insertion domain (Major DDE transposition pathways - Fig.7.3)) interacts with and assists in stabilizing a hairpin-loop structure at the transposon end formed as an intermediate during transposition[64][65][66] and therefore performs a crucial function. Another type of insertion domain is present at the same topological position in the eukaryotic Hermes transposase[52]. This long insertion domain is entirely α-helical. In contrast to Tn5 whose hairpin intermediate is formed on the transposon end, Hermes transposes using an intermediate in which a hairpin is formed on the flanking donor DNA[67][68]. It appears that the insertion domain assists stabilization of the Hermes hairpin intermediate. A similar α-helical insertion has been identified in the VDJ recombinase, RAG1[67] which also generates a hairpin intermediate similar to that of Hermes. In Hermes, the domain also assists in forming transposase multimers.
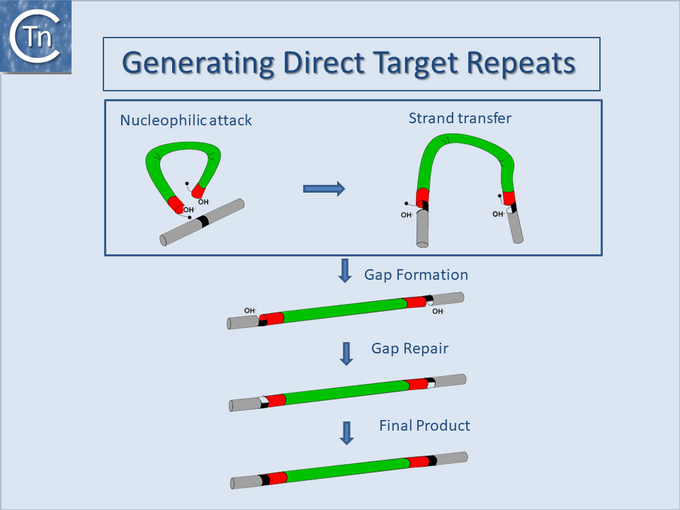
Direct target repeats
Another general feature of IS elements is that, on insertion, most generate short directly repeated sequences (DR) of the target DNA flanking the IS. The attack of each DNA strand at the target site by one of the two transposon ends in a staggered way during insertion provides an explanation for this observation. The DR is generated by repair of the “gap” between cleavage sites (Fig.18.5).
The length of the DR, generally between 2 and 14 bp, is characteristic for a given element and a given element will generally generate a duplication of fixed length. This is determined by the architecture of the transposition complex or transpososome which imposes constraints on the distance between cleavages on each strand of the target DNA[69][63][70].
However, certain ISs have been shown to generate DRs of atypical length at a low frequency, presumably reflecting small variations in the geometry of the transpososome (see [71]). Although some notable exceptions exist in which there is a systematic absence of DRs (either within a given family or in several independent transposition events of a given element), care should be taken in interpreting the absence of DRs in isolated cases.
A lack of DRs can simply result from homologous inter- or intra-molecular recombination between two IS elements, each with a different DR sequence. This would result in a hybrid element carrying one DR of each parent. It can also arise from the formation of adjacent deletions resulting from duplicative intramolecular transposition. In this case, a single copy of the DR is located on each of the reciprocal deletion products (see for example [72][73] or, more recently in a clinical context, [74].
Three IS, IS1549, IS1634 and IS1630, have been identified which appear to generate long DRs of quite variable length[75][76][77]. Two, IS1549 and IS1634, are distantly related to the IS4 family and one, IS1630, belongs to the IS30 family. The mechanism involved in generating such long DRs is at present unknown. However, it seems reasonable to propose that the target DNA may be able to form a loop within the transpososome and thereby bringing somewhat distant phophodiester bonds into close proximity.
Bibliography
- ↑ Derbyshire KM, Grindley ND . Cis preference of the IS903 transposase is mediated by a combination of transposase instability and inefficient translation. - Mol Microbiol: 1996 Sep, 21(6);1261-72 [PubMed:8898394] [DOI]
- ↑ Derbyshire KM, Kramer M, Grindley ND . Role of instability in the cis action of the insertion sequence IS903 transposase. - Proc Natl Acad Sci U S A: 1990 Jun, 87(11);4048-52 [PubMed:2161528] [DOI]
- ↑ Huisman O, Errada PR, Signon L, Kleckner N . Mutational analysis of IS10's outside end. - EMBO J: 1989 Jul, 8(7);2101-9 [PubMed:2551675] [DOI]
- ↑ Johnson RC, Reznikoff WS . DNA sequences at the ends of transposon Tn5 required for transposition. - Nature: 1983 Jul 21-27, 304(5923);280-2 [PubMed:6306482] [DOI]
- ↑ Makris JC, Nordmann PL, Reznikoff WS . Mutational analysis of insertion sequence 50 (IS50) and transposon 5 (Tn5) ends. - Proc Natl Acad Sci U S A: 1988 Apr, 85(7);2224-8 [PubMed:2832849] [DOI]
- ↑ Zerbib D, Prentki P, Gamas P, Freund E, Galas DJ, Chandler M . Functional organization of the ends of IS 1: specific binding site for an IS1-encoded protein. - Mol Microbiol: 1990 Sep, 4(9);1477-1486 [PubMed:28776821] [DOI]
- ↑ 7.0 7.1 7.2 Normand C, Duval-Valentin G, Haren L, Chandler M . The terminal inverted repeats of IS911: requirements for synaptic complex assembly and activity. - J Mol Biol: 2001 May 18, 308(5);853-71 [PubMed:11352577] [DOI]
- ↑ Ichikawa H, Ikeda K, Amemura J, Ohtsubo E . Two domains in the terminal inverted-repeat sequence of transposon Tn3. - Gene: 1990 Jan 31, 86(1);11-7 [PubMed:2155858] [DOI]
- ↑ May EW, Grindley ND . A functional analysis of the inverted repeat of the gamma delta transposable element. - J Mol Biol: 1995 Apr 7, 247(4);578-87 [PubMed:7723015] [DOI]
- ↑ Craigie R, Mizuuchi M, Mizuuchi K . Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. - Cell: 1984 Dec, 39(2 Pt 1);387-94 [PubMed:6094016] [DOI]
- ↑ Craig NL . Transposon Tn7. - Curr Top Microbiol Immunol: 1996, 204;27-48 [PubMed:8556868] [DOI]
- ↑ Rowland SJ, Dyke KG . Tn552, a novel transposable element from Staphylococcus aureus. - Mol Microbiol: 1990 Jun, 4(6);961-75 [PubMed:2170815] [DOI]
- ↑ Rowland SJ, Sherratt DJ, Stark WM, Boocock MR . Tn552 transposase purification and in vitro activities. - EMBO J: 1995 Jan 3, 14(1);196-205 [PubMed:7828593] [DOI]
- ↑ Gierl A . The En/Spm transposable element of maize. - Curr Top Microbiol Immunol: 1996, 204;145-59 [PubMed:8556865] [DOI]
- ↑ Kunze R . The maize transposable element activator (Ac). - Curr Top Microbiol Immunol: 1996, 204;161-94 [PubMed:8556866] [DOI]
- ↑ Berger N, Heller AE, Störmann KD, Pfaff E . Characterization of chimeric enzymes between caprine arthritis--encephalitis virus, maedi--visna virus and human immunodeficiency virus type 1 integrases expressed in Escherichia coli. - J Gen Virol: 2001 Jan, 82(Pt 1);139-148 [PubMed:11125167] [DOI]
- ↑ Mahillon J, Chandler M . Insertion sequences. - Microbiol Mol Biol Rev: 1998 Sep, 62(3);725-74 [PubMed:9729608] [DOI]
- ↑ Machida C, Machida Y . Regulation of IS1 transposition by the insA gene product. - J Mol Biol: 1989 Aug 20, 208(4);567-74 [PubMed:2553980] [DOI]
- ↑ Zerbib D, Polard P, Escoubas JM, Galas D, Chandler M . The regulatory role of the IS1-encoded InsA protein in transposition. - Mol Microbiol: 1990 Mar, 4(3);471-7 [PubMed:2162466] [DOI]
- ↑ Stalder R, Caspers P, Olasz F, Arber W . The N-terminal domain of the insertion sequence 30 transposase interacts specifically with the terminal inverted repeats of the element. - J Biol Chem: 1990 Mar 5, 265(7);3757-62 [PubMed:2154486]
- ↑ Lavoie BD, Chaconas G . Transposition of phage Mu DNA. - Curr Top Microbiol Immunol: 1996, 204;83-102 [PubMed:8556870] [DOI]
- ↑ Lavoie BD, Shaw GS, Millner A, Chaconas G . Anatomy of a flexer-DNA complex inside a higher-order transposition intermediate. - Cell: 1996 May 31, 85(5);761-71 [PubMed:8646783] [DOI]
- ↑ Maekawa T, Amemura-Maekawa J, Ohtsubo E . DNA binding domains in Tn3 transposase. - Mol Gen Genet: 1993 Jan, 236(2-3);267-74 [PubMed:8382339] [DOI]
- ↑ Maekawa T, Ohtsubo E . Identification of the region that determines the specificity of binding of the transposases encoded by Tn3 and gamma delta to the terminal inverted repeat sequences. - Jpn J Genet: 1994 Jun, 69(3);269-85 [PubMed:8080658] [DOI]
- ↑ Wiegand TW, Reznikoff WS . Interaction of Tn5 transposase with the transposon termini. - J Mol Biol: 1994 Jan 14, 235(2);486-95 [PubMed:8289277] [DOI]
- ↑ Tavakoli NP, DeVost J, Derbyshire KM . Defining functional regions of the IS903 transposase. - J Mol Biol: 1997 Dec 12, 274(4);491-504 [PubMed:9417930] [DOI]
- ↑ Polard P, Ton-Hoang B, Haren L, Bétermier M, Walczak R, Chandler M . IS911-mediated transpositional recombination in vitro. - J Mol Biol: 1996 Nov 22, 264(1);68-81 [PubMed:8950268] [DOI]
- ↑ 28.0 28.1 Haren L, Ton-Hoang B, Chandler M . Integrating DNA: transposases and retroviral integrases. - Annu Rev Microbiol: 1999, 53;245-81 [PubMed:10547692] [DOI]
- ↑ Weinreich MD, Mahnke-Braam L, Reznikoff WS . A functional analysis of the Tn5 transposase. Identification of domains required for DNA binding and multimerization. - J Mol Biol: 1994 Aug 12, 241(2);166-77 [PubMed:8057357] [DOI]
- ↑ Jain C, Kleckner N . Preferential cis action of IS10 transposase depends upon its mode of synthesis. - Mol Microbiol: 1993 Jul, 9(2);249-60 [PubMed:8412678] [DOI]
- ↑ 31.0 31.1 31.2 Haren L, Polard P, Ton-Hoang B, Chandler M . Multiple oligomerisation domains in the IS911 transposase: a leucine zipper motif is essential for activity. - J Mol Biol: 1998, 283(1);29-41 [PubMed:9761671] [DOI]
- ↑ Duval-Valentin G, Chandler M . Cotranslational control of DNA transposition: a window of opportunity. - Mol Cell: 2011 Dec 23, 44(6);989-96 [PubMed:22195971] [DOI]
- ↑ Buchner JM, Robertson AE, Poynter DJ, Denniston SS, Karls AC . Piv site-specific invertase requires a DEDD motif analogous to the catalytic center of the RuvC Holliday junction resolvases. - J Bacteriol: 2005 May, 187(10);3431-7 [PubMed:15866929] [DOI]
- ↑ Choi S, Ohta S, Ohtsubo E . A novel IS element, IS621, of the IS110/IS492 family transposes to a specific site in repetitive extragenic palindromic sequences in Escherichia coli. - J Bacteriol: 2003 Aug, 185(16);4891-900 [PubMed:12897009] [DOI]
- ↑ 35.0 35.1 35.2 Haren L, Normand C, Polard P, Alazard R, Chandler M . IS911 transposition is regulated by protein-protein interactions via a leucine zipper motif. - J Mol Biol: 2000 Feb 25, 296(3);757-68 [PubMed:10677279] [DOI]
- ↑ Chaconas G, Lavoie BD, Watson MA . DNA transposition: jumping gene machine, some assembly required. - Curr Biol: 1996 Jul 1, 6(7);817-20 [PubMed:8805293] [DOI]
- ↑ Weinreich MD, Gasch A, Reznikoff WS . Evidence that the cis preference of the Tn5 transposase is caused by nonproductive multimerization. - Genes Dev: 1994 Oct 1, 8(19);2363-74 [PubMed:7958902] [DOI]
- ↑ Ronning DR, Guynet C, Ton-Hoang B, Perez ZN, Ghirlando R, Chandler M, Dyda F . Active site sharing and subterminal hairpin recognition in a new class of DNA transposases. - Mol Cell: 2005 Oct 7, 20(1);143-54 [PubMed:16209952] [DOI]
- ↑ Hickman AB, James JA, Barabas O, Pasternak C, Ton-Hoang B, Chandler M, Sommer S, Dyda F . DNA recognition and the precleavage state during single-stranded DNA transposition in D. radiodurans. - EMBO J: 2010 Nov 17, 29(22);3840-52 [PubMed:20890269] [DOI]
- ↑ Bolland S, Kleckner N . The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. - Cell: 1996 Jan 26, 84(2);223-33 [PubMed:8565068] [DOI]
- ↑ Katz RA, Skalka AM . The retroviral enzymes. - Annu Rev Biochem: 1994, 63;133-73 [PubMed:7526778] [DOI]
- ↑ Jones KS, Coleman J, Merkel GW, Laue TM, Skalka AM . Retroviral integrase functions as a multimer and can turn over catalytically. - J Biol Chem: 1992 Aug 15, 267(23);16037-40 [PubMed:1322888]
- ↑ 43.0 43.1 Bao KK, Wang H, Miller JK, Erie DA, Skalka AM, Wong I . Functional oligomeric state of avian sarcoma virus integrase. - J Biol Chem: 2003 Jan 10, 278(2);1323-7 [PubMed:12446721] [DOI]
- ↑ Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V . HIV-1 integrase crosslinked oligomers are active in vitro. - Nucleic Acids Res: 2005, 33(3);977-86 [PubMed:15718297] [DOI]
- ↑ Ren G, Gao K, Bushman FD, Yeager M . Single-particle image reconstruction of a tetramer of HIV integrase bound to DNA. - J Mol Biol: 2007 Feb 9, 366(1);286-94 [PubMed:17157316] [DOI]
- ↑ Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P . Structural basis for functional tetramerization of lentiviral integrase. - PLoS Pathog: 2009 Jul, 5(7);e1000515 [PubMed:19609359] [DOI]
- ↑ Michel F, Crucifix C, Granger F, Eiler S, Mouscadet JF, Korolev S, Agapkina J, Ziganshin R, Gottikh M, Nazabal A, Emiliani S, Benarous R, Moras D, Schultz P, Ruff M . Structural basis for HIV-1 DNA integration in the human genome, role of the LEDGF/P75 cofactor. - EMBO J: 2009 Apr 8, 28(7);980-91 [PubMed:19229293] [DOI]
- ↑ Tang M, Cecconi C, Bustamante C, Rio DC . Analysis of P element transposase protein-DNA interactions during the early stages of transposition. - J Biol Chem: 2007 Sep 28, 282(39);29002-29012 [PubMed:17644523] [DOI]
- ↑ Lohe AR, Sullivan DT, Hartl DL . Subunit interactions in the mariner transposase. - Genetics: 1996 Nov, 144(3);1087-95 [PubMed:8913752] [DOI]
- ↑ Richardson JM, Finnegan DJ, Walkinshaw MD . Crystallization of a Mos1 transposase-inverted-repeat DNA complex: biochemical and preliminary crystallographic analyses. - Acta Crystallogr Sect F Struct Biol Cryst Commun: 2007 May 1, 63(Pt 5);434-7 [PubMed:17565190] [DOI]
- ↑ Richardson JM, Colloms SD, Finnegan DJ, Walkinshaw MD . Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. - Cell: 2009 Sep 18, 138(6);1096-108 [PubMed:19766564] [DOI]
- ↑ 52.0 52.1 Hickman AB, Perez ZN, Zhou L, Musingarimi P, Ghirlando R, Hinshaw JE, Craig NL, Dyda F . Molecular architecture of a eukaryotic DNA transposase. - Nat Struct Mol Biol: 2005 Aug, 12(8);715-21 [PubMed:16041385] [DOI]
- ↑ Perez ZN, Musingarimi P, Craig NL, Dyda F, Hickman AB . Purification, crystallization and preliminary crystallographic analysis of the Hermes transposase. - Acta Crystallogr Sect F Struct Biol Cryst Commun: 2005 Jun 1, 61(Pt 6);587-90 [PubMed:16511103] [DOI]
- ↑ Hickman AB, Ewis HE, Li X, Knapp JA, Laver T, Doss AL, Tolun G, Steven AC, Grishaev A, Bax A, Atkinson PW, Craig NL, Dyda F . Structural basis of hAT transposon end recognition by Hermes, an octameric DNA transposase from Musca domestica. - Cell: 2014 Jul 17, 158(2);353-367 [PubMed:25036632] [DOI]
- ↑ Rice P, Craigie R, Davies DR . Retroviral integrases and their cousins. - Curr Opin Struct Biol: 1996 Feb, 6(1);76-83 [PubMed:8696976] [DOI]
- ↑ Grindley ND, Leschziner AE . DNA transposition: from a black box to a color monitor. - Cell: 1995 Dec 29, 83(7);1063-6 [PubMed:8548793] [DOI]
- ↑ Hu JC, O'Shea EK, Kim PS, Sauer RT . Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. - Science: 1990 Dec 7, 250(4986);1400-3 [PubMed:2147779] [DOI]
- ↑ Andrews JD, Gloor GB . A role for the KP leucine zipper in regulating P element transposition in Drosophila melanogaster. - Genetics: 1995 Oct, 141(2);587-94 [PubMed:8647395] [DOI]
- ↑ Lewis LA, Astatke M, Umekubo PT, Alvi S, Saby R, Afrose J . Soluble expression, purification and characterization of the full length IS2 Transposase. - Mob DNA: 2011 Oct 27, 2;14 [PubMed:22032517] [DOI]
- ↑ Ivics Z, Izsvak Z, Minter A, Hackett PB . Identification of functional domains and evolution of Tc1-like transposable elements. - Proc Natl Acad Sci U S A: 1996 May 14, 93(10);5008-13 [PubMed:8643520] [DOI]
- ↑ Lysnyansky I, Calcutt MJ, Ben-Barak I, Ron Y, Levisohn S, Methé BA, Yogev D . Molecular characterization of newly identified IS3, IS4 and IS30 insertion sequence-like elements in Mycoplasma bovis and their possible roles in genome plasticity. - FEMS Microbiol Lett: 2009 May, 294(2);172-82 [PubMed:19416360] [DOI]
- ↑ Utsumi R, Ikeda M, Horie T, Yamamoto M, Ichihara A, Taniguchi Y, Hashimoto R, Tanabe H, Obata K, Noda M . Isolation and characterization of the IS3-like element from Thermus aquaticus. - Biosci Biotechnol Biochem: 1995 Sep, 59(9);1707-11 [PubMed:8520113] [DOI]
- ↑ 63.0 63.1 Dyda F, Chandler M, Hickman AB . The emerging diversity of transpososome architectures. - Q Rev Biophys: 2012 Nov, 45(4);493-521 [PubMed:23217365] [DOI]
- ↑ Davies DR, Goryshin IY, Reznikoff WS, Rayment I . Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. - Science: 2000 Jul 7, 289(5476);77-85 [PubMed:10884228] [DOI]
- ↑ Steiniger-White M, Rayment I, Reznikoff WS . Structure/function insights into Tn5 transposition. - Curr Opin Struct Biol: 2004 Feb, 14(1);50-7 [PubMed:15102449] [DOI]
- ↑ Steiniger M, Metzler J, Reznikoff WS . Mutation of Tn5 transposase beta-loop residues affects all steps of Tn5 transposition: the role of conformational changes in Tn5 transposition. - Biochemistry: 2006 Dec 26, 45(51);15552-62 [PubMed:17176076] [DOI]
- ↑ 67.0 67.1 Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL . Transposition of hAT elements links transposable elements and V(D)J recombination. - Nature: 2004 Dec 23, 432(7020);995-1001 [PubMed:15616554] [DOI]
- ↑ Hickman AB, Voth AR, Ewis H, Li X, Craig NL, Dyda F . Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase. - Nucleic Acids Res: 2018 Nov 2, 46(19);10286-10301 [PubMed:30239795] [DOI]
- ↑ Gueguen E, Rousseau P, Duval-Valentin G, Chandler M . The transpososome: control of transposition at the level of catalysis. - Trends Microbiol: 2005 Nov, 13(11);543-9 [PubMed:16181782] [DOI]
- ↑ Montaño SP, Rice PA . Moving DNA around: DNA transposition and retroviral integration. - Curr Opin Struct Biol: 2011 Jun, 21(3);370-8 [PubMed:21439812] [DOI]
- ↑ Hickman AB, Dyda F . Mechanisms of DNA Transposition. - Microbiol Spectr: 2015 Apr, 3(2);MDNA3-0034-2014 [PubMed:26104718] [DOI]
- ↑ Weinert TA, Schaus NA, Grindley ND . Insertion sequence duplication in transpositional recombination. - Science: 1983 Nov 18, 222(4625);755-65 [PubMed:6314502] [DOI]
- ↑ Turlan C, Chandler M . IS1-mediated intramolecular rearrangements: formation of excised transposon circles and replicative deletions. - EMBO J: 1995 Nov 1, 14(21);5410-21 [PubMed:7489730] [DOI]
- ↑ He S, Hickman AB, Varani AM, Siguier P, Chandler M, Dekker JP, Dyda F . Insertion Sequence IS26 Reorganizes Plasmids in Clinically Isolated Multidrug-Resistant Bacteria by Replicative Transposition. - mBio: 2015 Jun 9, 6(3);e00762 [PubMed:26060276] [DOI]
- ↑ Calcutt MJ, Lavrrar JL, Wise KS . IS1630 of Mycoplasma fermentans, a novel IS30-type insertion element that targets and duplicates inverted repeats of variable length and sequence during insertion. - J Bacteriol: 1999 Dec, 181(24);7597-607 [PubMed:10601219] [DOI]
- ↑ Plikaytis BB, Crawford JT, Shinnick TM . IS1549 from Mycobacterium smegmatis forms long direct repeats upon insertion. - J Bacteriol: 1998 Mar, 180(5);1037-43 [PubMed:9495740] [DOI]
- ↑ Vilei EM, Nicolet J, Frey J . IS1634, a novel insertion element creating long, variable-length direct repeats which is specific for Mycoplasma mycoides subsp. mycoides small-colony type. - J Bacteriol: 1999 Feb, 181(4);1319-23 [PubMed:9973360] [DOI]