IS Families/IS4 and related families
Contents
- 1 General
- 2 IS4 family
- 3 IS10 and IS50
- 3.1 Compound transposons
- 3.2 Overall Organization and Terminal inverted repeats
- 3.3 Regulation of transposition activity
- 3.4 Dam methylation
- 3.5 Small RNAs
- 3.6 Host proteins: IHF
- 3.7 Host proteins: HNS
- 3.8 Host proteins: Hfq
- 3.9 Other host proteins
- 3.10 Transposase organization
- 3.11 Mechanism
- 3.12 Protein structure and the transpososome
- 4 IS231
- 5 IS701 family
- 6 ISH3 family
- 7 IS1634 family
- 8 Acknowledgements
- 9 Bibliography
General
The accumulation of additional related IS has permitted a more detailed analysis of the IS4 family which had already become heterogeneous displaying extremely elevated levels of internal divergence[1]. Although this family remains extremely heterogeneous, members form coherent subgroups or clusters. Each subgroup is related to others through members with low level BLAST scores and no members of other families are found at intervening positions. Based on more than 200 IS4-related sequences from bacterial and archaeal genome sequences, seven subgroups (IS10, IS231, IS4, IS4Sa, IS50, ISH8 and ISPerp1) and three families (IS701, ISH3 and IS1634) were defined. Separation into three families (Table Characteristics of IS families; Fig.4.2) was principally due to variations in an important conserved YREK motif, a division which is supported by the IR sequences and the associated DR (Fig.IS4.1 and Figs. IS4.1.2, IS4.1.3, IS4.1.4, IS4.1.5, IS4.1.6, IS4.1.7, IS4.1.8, IS4.1.9, and IS4.1.10 in slideshow format below).
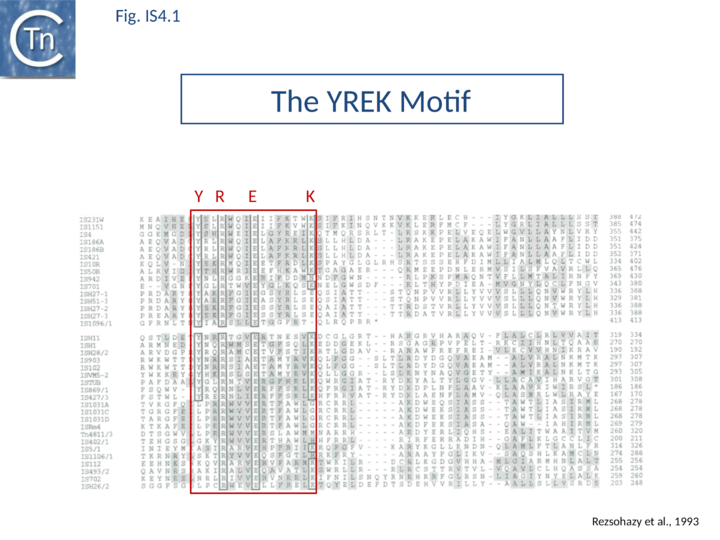
Members encode a Tpase with an insertion domain rich in β-strand and located between the second D and the E of the DDE motif[2] (Fig.7.3). That of the IS50 Tpase[3] is the only example which has structural support[4] although bioinformatic analysis[2] indicated that ISH3 (e.g. ISC1359 and ISC1439), IS701 (e.g. IS701 and ISRso17[5]), and IS1634 (family members e.g. IS1634, ISMac5, ISPlu4[6]) also exhibit a similar insertion domain (Table Transposases examined by secondary structure prediction programs).
IS4 family
The IS4 family originally included a diverse collection of IS characterized by three conserved domains in the Tpase [N2, N3 and C1 containing D, D and E, respectively[3]]. In addition to the conserved DDE triad, this family is defined by the presence of an additional tetrad YREK[1]. For IS50, this is involved in coordination of a terminal phosphate group at the 5’ end of the cleaved IS [7][8].
IS10 and IS50
Compound transposons
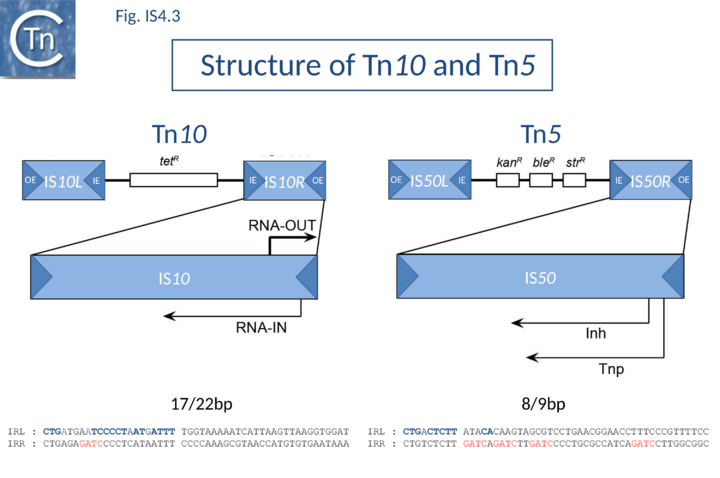
IS10, which forms part of the composite tetracycline resistance transposon Tn10, and IS50, which forms part of the kanamycin resistance transposon Tn5, are certainly the best characterized members of this group (Fig.IS4.3).
Tn5 and Tn10 have been extensively reviewed[8][9][10][11] and the entire nucleotide sequence of Tn10 is available[12].
In both cases, the flanking IS are in an inverted configuration, and the activity of one of the two flanking IS is compromised by inactivating mutations. This presumably stabilizes the transposon since it reduces the autonomy of one of the two IS copies.
For Tn5, the Inside End, IE (proximal to the resistance gene), has undergone a mutation which introduces a premature termination codon into the transposase gene and creates a promoter to drive the kanamycin resistance[13][14][15].
Overall Organization and Terminal inverted repeats
IS10 and IS50 Tpases are expressed from a single long reading frame by convention shown as expressed from left to right and bordered by short inverted terminal repeats with a typical two-domain organization (Fig.18.2).
In both Tn5 and Tn10, the ends of the individual flanking IS are called outside (OE = IRL) and inside (IE = IRR) ends to describe their relative position in the Tn10 and Tn5 compound transposons. IS10 carries 22 bp terminal IRs and between 13 and 27 base pairs of each IR are absolutely required although sequences up to 70 at each end can influence transposition[16]. Moreover, IE and OE are not equivalent. OE includes a binding site for IHF (which intervenes in formation of the transpososome) whereas IE does not[17].
The ends of IS50 are shorter but the terminal 19bp are critical for transposition[18]. As for IS10, OE and IE are not equivalent [13]. The crystal structure of the Tn5 transposase/IR complex revealed that almost all of the 19 bp are contacted by protein[4] (Fig.23.3).
The observation that OE and IE are not equivalent implies subtle differences in transposition dynamics of the entire Tn10 compared to IS10 alone.
Regulation of transposition activity
The elements exhibit an elaborate ensemble of mechanisms to control their activity and are protected from activation by impinging external transcription by an inverted repeat sequence located close to the left end[19][20][21]. Activity is also regulated by various host DNA architectural proteins such as IHF and H-NS[8].
Dam methylation
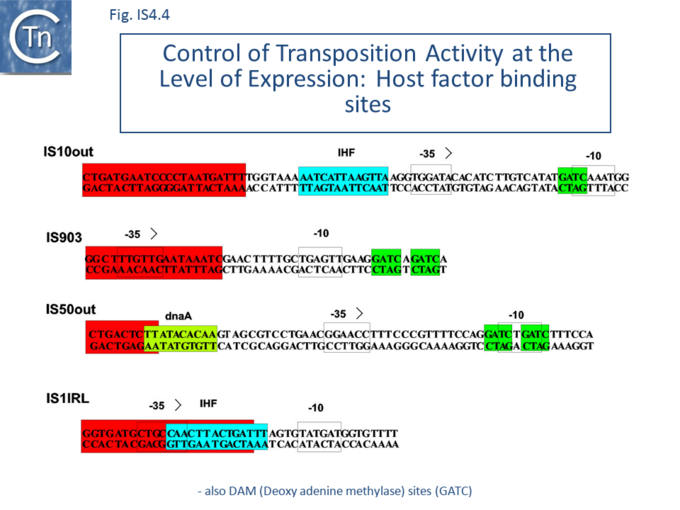
IS10 contains a Dam methylation site in the -10 hexamer of the Tpase promoter (pIN) (Fig.2.3; Fig.IS4.4, and Fig.IS4.5). Following replication, this site becomes transiently hemi-methylated and promoter strength is increased. Dam methylation sites within the Tpase promoter, P1, of IS50 play a similar role.
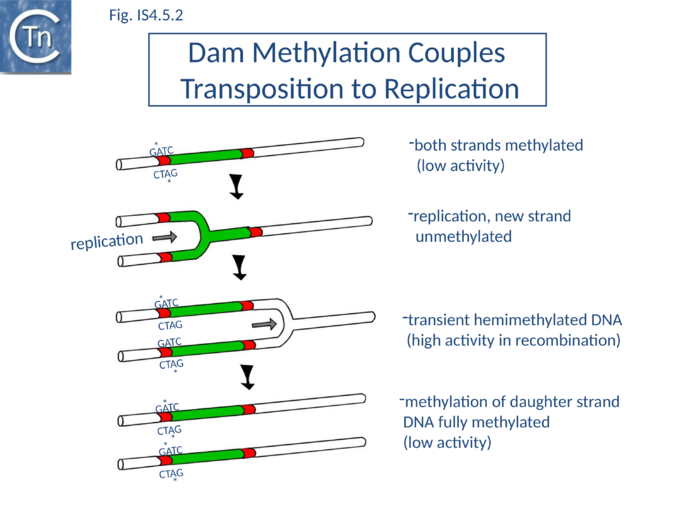
Dam methylation also exerts control at another level. In addition to the Dam site located in IRL, another site is localized in IRR of IS10. Fully methylated sites cause reduced IR activity whereas hemimethylated IRs exhibit increased activity. With this arrangement, both Tpase expression and transposition activity are coupled to the replication of the donor molecule (Fig.IS4.5). Since transposition of IS10 is non-replicative, this assures that passive replication of the IS occurs before transposition takes place.
For IS50, transposition activity is reduced by methylation of three consecutive Dam methylation sites located in IE[22] and has been directly attributed to interference with Tnp binding[9].
Small RNAs
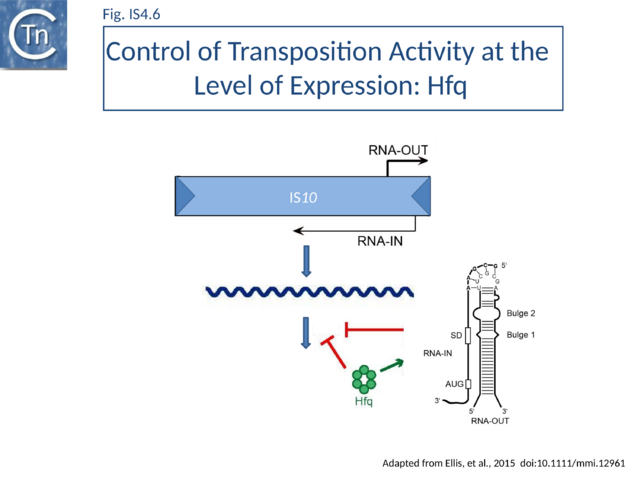
Tn10 encodes an antisense RNA (RNA-OUT) perfectly complementary to 35 nucleotides of the transposase mRNA (RNA-IN)[23][24][25][26] (Fig.IS4.6). IS10 Tpase expression is controlled in trans by RNA-OUT which is transcribed from an outward directed promoter located proximal to IRL (pOUT). This RNA, RNA-OUT, is perfectly complementary to 35 nucleotides of transposase mRNA (RNA-IN) and pairs with RNA-IN to occlude the ribosome binding site and inhibit ribosome binding. This inhibits transposase translation and decreases its stability, thereby acting as a potent negative transposition regulator. At the time of its deiscovery, RNA-OUT was only the second example of an anti-RNA. The first to be identified was “RNAI”, involved in regulation of replication of plasmid ColE1[27][28].
In IS50, control in trans is exerted by a second protein, the inhibitor protein, Inh. This is translated in the same frame as transposase, Tnp, but uses an alternative initiation codon and lacks the N-terminal 55 amino acids. It probably employs a separate (and possibly competing) promoter, P2, whose activity is not affected by Dam methylation. Both P1 and P2 are located downstream from the terminal IR[21] (Fig.IS4.3). It is thought that the inhibitory action of Inh involves the formation of (inactive) heteromultimers between Inh and Tnp.
Host proteins: IHF
The host IHF (Integration Host Factor, first identified as a requirement for bacteriophage l integration and excision; (see [29]) protein binds within the left end of IS10, interior to the IRL sequence, and subtly influences the nature of transposition products[30][31]. IHF appears to facilitate formation of the IS10 transpososome[31]. Binding to IS10 OE (left end) produces an 180° bend in the DNA resulting in transposase contacts with both terminal and sub-terminal regions of the IR[32] (Fig.IS4.4).
Host proteins: HNS
H-NS has also been implicated in IS10 transposition in vivo[33] (Fig.IS4.7). This was thought to involve target capture since an excised transposon fragment, a precursor to target capture, accumulated in in vivo induction assays in the absence of H-NS.
H-NS is a highly expressed nucleoid binding protein, widely distributed in enterobacteria, and often acts as a transcriptional repressor. HNS has structure-specific DNA binding activity. It preferentially binds A-T rich sequences and is sensitive to the shape of the minor groove of DNA[34] (see also [35]).
H-NS binds to and stabilizes the folded transpososome and stimulates strand transfer in a full transposition reaction[36][37][38]. It appears to recognize a distorted DNA structure in the flanking donor DNA within the transpososome. Footprinting indicated that H-NS bound within the outer IS10 end (OE) close to the IHF binding site. The crosslinking analysis suggested that H-NS recognizes both structural features in the transpososome and the transposase protein.
For IS50, genetic studies revealed a 6-fold drop in transposition for a Tn5 derivative under conditions of hns deficiency suggesting a positive regulatory role[39]. Footprinting and mutational analyses showed that H-NS binds Tn5 transpososomes with high specificity with three potential binding sites within the terminal 20 bp of the transposon end and crosslinking also revealed that H-NS could directly contact transposase.
The IS10 transpososome undergoes clear changes in configuration during the course of transposition[8] [32][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56]. The role of both DNA architectural proteins IHF and H-NS is to assure correct bending of the DNA within the transpososome. H-NS may function in IS50 transpososome assembly based on the observation that H-NS can both promote transpososome formation and bind to a single end-transposase complex in vitro[39].
Host proteins: Hfq
Although Hfq is involved in H-NS expression[57], in Tn10 it acts independently of H-NS to down-regulate transposition by regulating transposase expression[58].
There is an increase in transposase expression in an hfq mutant but required a context in which the reporter gene used included the native transposase promoter and sequences required for translational control. This is consistent with Hfq acting as a post-transcriptional regulator of transposase expression[58].
Hfq typically functions in ribo-regulation by aiding in the pairing of RNA species. Hfq might therefore play a role in the antisense pairing system of Tn10. It was demonstrated that in vitro Hfq binds both RNA-IN and RNA-OUT and accelerates the rate of pairing (Fig.IS4.6).
Hfq can also function independently of the antisense system to down-regulate Tn10/IS10 transposition, presumably by its capacity to bind directly to RNA-IN[58]. RNase footprinting studies were consistent with the idea that Hfq binds directly to the translation initiation region of RNA-IN[59].
RNA-OUT is highly structured and, although it has 35 nts of perfect complementarity with RNA-IN, the capacity to form a stable hybrid is probably limited.
RNase footprinting of Hfq-RNA-OUT complexes revealed that Hfq destabilized the stem of the RNA-OUT stem-loop structure Ross[60]. Based on genetic studies it had been presumed that discontinuities in the RNA-OUT stem would be sufficient to destabilize the stem for full pairing[24].
RNase footprinting and EMSA[60] has provided detailed information concerning Hfq interactions with RNA-IN and RNA-OUT. In particular this has identified three high affinity binding sites in RNA-IN and one in RNA-OUT. Studies with mutant Hfq derivatives has led to a model for Hfq function in promoting pairing[61].
For IS50, Hfq inhibits Tn5 transposase expression at the transcriptional level. Transposition of an E. coli chromosomal Tn5 copy increased about 10-fold under conditions of hfq deficiency. Unlike Tn10, an ‘up-expression’ phenotype was observed in both transcriptional and translation fusion reporter genes. Hfq probably negatively regulates transposase gene transcription. Indeed the steady-state level of transposase transcript increased about 3-fold in conditions of hfq deficiency [8], an observation which should be explored in more detail.
Other host proteins
Binding sites for additional host proteins occur in IS50. OE includes a binding site for the host DnaA protein whereas IE carries a binding site for the host protein, Fis. Transposition activity is reduced in a dnaA host and by the presence of the Fis site[62] (Fig.IS4.4).
Transposase organization
There has been extensive functional analysis of the Tpases (or derivatives) of both elements by partial proteolysis and mutagenesis[63][64][65][66][67].
There are three distinct domains within both the IS50 and IS10 transposases (for DNA binding, multimerization and catalysis)[66] and residues contributed by all of them participate in DNA binding. IS50 produces two proteins, the transposase, and an inhibitor protein (Inh) which is the product of an alternative internal transcript. The IS50 transcriptional start sites responsible for transposase expression and transposition inhibitor protein expression, and the likely translational start site of transposase was determined by N-terminal analysis. It starts at position 93 of IS50. Three in vivo transcripts T1, T2, and T3, initiate in this region. Only T1 starts upstream from the transposase N-terminus. The three transcripts initiate at independent but overlapping promoters near the left IS50 end. T1 encodes transposase while T2 is largely responsible for inhibitor protein expression. The T3 coding capacity is not known[21].
Mechanism
Both IS10 and IS50 transpose by a "cut-and-paste" mechanism. The mechanism of transposition of other members of this group is assumed to be similar but is at present unknown.
IS10 and IS50 were the first IS for which cell-free in vitro systems were developed[68][69][70].
IS10 and IS50 use a similar, if not identical, transposition mechanism (Fig.IS4.8).
Cut-and-paste transposition of IS10 was elegantly demonstrated genetically[71] and biochemically[65] several decades ago.
Tn10 is excised of from the donor site during transposition by flush double-strand cleavages at the transposon termini[72][73] and cleavage of both strands at one transposon end occurs in a specific order. Cleavage of the transferred strand is followed by cleavage of the non-transferred strand[74] and involves repeated use of only a single active site[75].
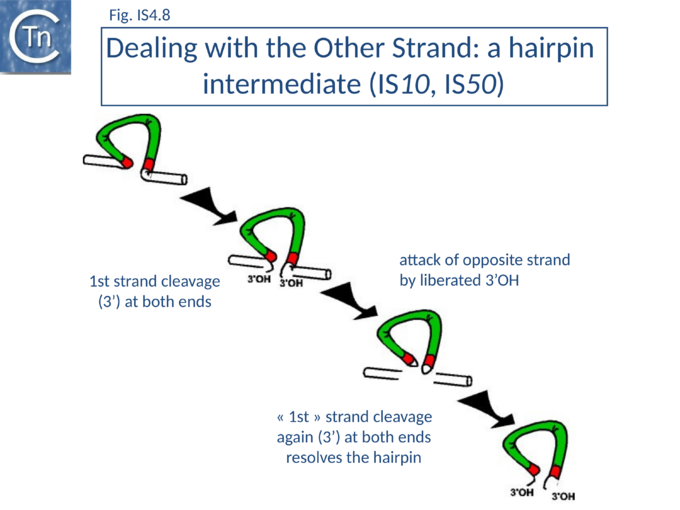
It was later shown that both IS10 and IS50 are excised via a hairpin intermediate in which the complementary strands at each transposon end are covalently joined[76][77] (Fig.IS4.8).
A 3’OH generated by cleavage of the transferred DNA strand attacks the opposite (complementary) strand to generate the hairpin (in which both strands are joined at the transposon end). Transposase then resolves this structure by hydrolysis of the bridged hairpin to regenerate the 3’OH of the transferred strand liberating the transposon from flanking donor DNA. This intermediate which is thought to be maintained in a non-covalently joined circular form by the transposase, proceeds to strand transfer into a suitable target molecule. This excision model explains how a single molecule of transposase with a single active site can make a flush double-strand break in a DNA molecule to release transposon from flanking donor DNA sequences[77][78]. Equivalent observations have been made for Tn5[76].
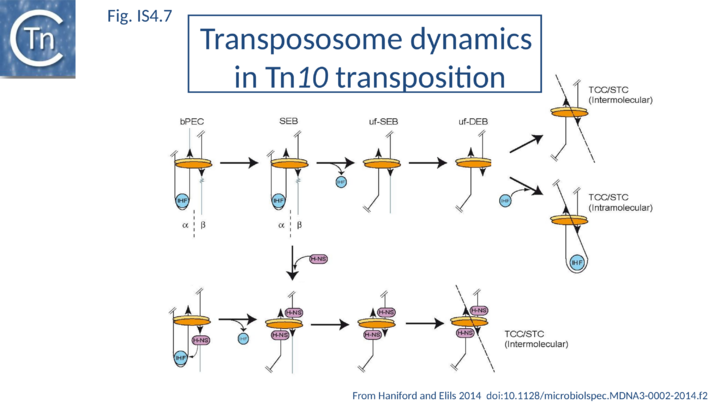
The different steps involved in IS10 transposition have been exquisitely detailed using EMSA and other biochemical approaches[8][32] [33][36][37][38][36] [39][40][41][42][43][44][45][79] [51][52][53][54][55] [56][80][81] (Fig.IS4.7). This involves the idea of a molecular spring implicating protein-mediated folding and unfolding of the transpososome[32]. In a first step, a transposase dimer is thought to bind two antiparallel IS10 ends (defined as α and β) with assistance from IHF (by its capacity to bend the one end, the α-arm) to form a bPEC (paired end complex). This then undergoes cleavage in which flanking donor DNA is cleaved from one end while maintaining the folded α-arm to generate a single end break complex (αSEB). IHF is then lost to generate an unfolded single end break complex (uf-SEB). The second end is then cleaved to liberate the second donor DNA flank to generate an unfolded double end break complex (uf-DEB). There is then a choice: either uf-DEB can capture a target DNA (TCC – target capture complex) and catalyze intermolecular strand transfer Strand Transfer Complex, STC) or IHF can re-bind and re-fold a transposon arm which promotes intramolecular target capture and strand transfer where part of the transposon serves as the target DNA. Transpososome folding and unfolding is key to determining the relative frequency of inter- and intra-molecular transposition.
H-NS also appears to impact IS10 transpososome dynamics. It forms dimers in solution and higher-order oligomers on DNA, binds non-specifically with low affinity but with high affinity in a structure-specific way[82]. An H-NS dimer is thought to bind the DNA flank of the folded α-SEB on the arm not bound by IHF, the α-arm. IHF and H-NS compete for the same site(s) and H-NS stabilizes forms of the unfolded transpososome and facilitates displacement of IHF. This allows the α-arm to unfold and recruit additional H-NS dimers. H-NS binding within the transposon helps to maintain the transpososome in an unfolded state stabilizing the fully cleaved, unfolded uf-DEB and promoting intermolecular target capture. It is not clear whether H-NS interacts initially with bPEC and continues through α-SEB, uf-SEB and finally uf-DEB, “channeling” the transpososome to intermolecular targets. The idea of channeling was initially introduced to explain the effect of IHF-promoted folding on inter- and intramolecular transposition[31][68].
Protein structure and the transpososome
Although the biochemical and genetic analysis of the IS10 transpososome has yielded a detailed model of its assembly, there is no structural information available.
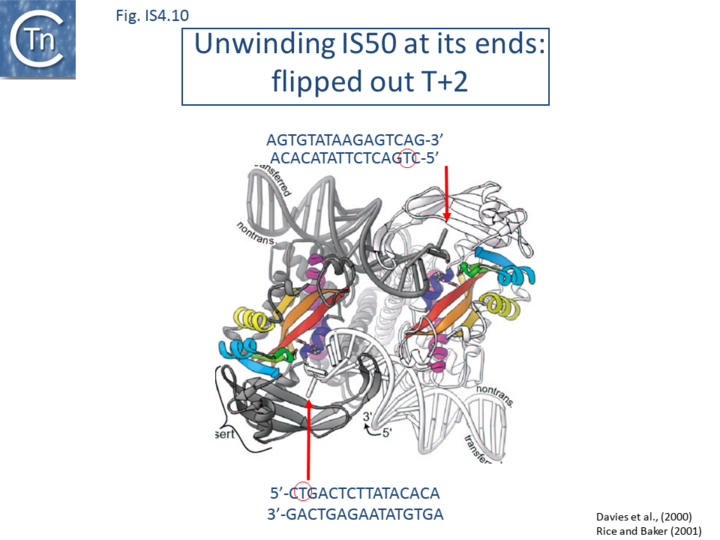
However, the structures of both the IS50 inhibitor, Inh,[83] and of the Tpase complex with the terminal IRs have been determined[4] (Fig.IS4.9 and Fig.IS4.10). The structure of the IS50 transpososome was the first to be elucidated. The complex structure indicates that the two transposon ends are aligned in an antiparallel configuration by two transposase monomers. This has provided a structural basis for the observation that end cleavage occurs in trans (i.e. that Tpase bound at one end catalyzes the cleavage of the opposite end[84]). These studies have provided a detailed picture of the IS50 transposition mechanism at the chemical level.
Uncomplexed IS50 transposase is a monomer[85] and the crystal structure shows how DNA binding and multimerization are inextricably linked.
The N-terminal sequence-specific DNA binding domain of one IS50 transposase monomer recognizes the internal IR region between nts 5 and 16. Formation of the initial subterminal ‘cis’ DNA contacts is thought to induce a conformational change affecting the C-terminal domain, creating an interface for dimerization[85]. Upon dimerization, the transposon tip (and hence the cleavage site) is inserted into the active site of the other monomer forming a set of ‘trans’ contacts.
The IS50 transposase N-terminal DNA-binding domain and the C-terminal dimerization domain inhibit each other’s activity. Full-length transposase appears not to bind its transposon end as a monomer. However, a truncated protein deleted for a C-terminal dimerization domain does so readily[86]. However, removal of the N-terminal DNA-binding domain allows the truncated protein to dimerize[85], whereas the full-length transposase does not. A number of conformational rearrangements must, therefore, occur during transpososome assembly to relieve these reciprocal inhibitions.
Presumably, as for IS911, this is the result of co-translational binding of the nascent transposase[87] and contributes to the ‘cis’ activity of the transposases[70][88].
In the case of IS10, a short treatment with alcohol served to activate transposase in an in vitro system[69] suggesting that full length protein must be partially denatured to function. Additionally it was shown that two transposase proteolytic fragments retained transposition activity in vitro [66].
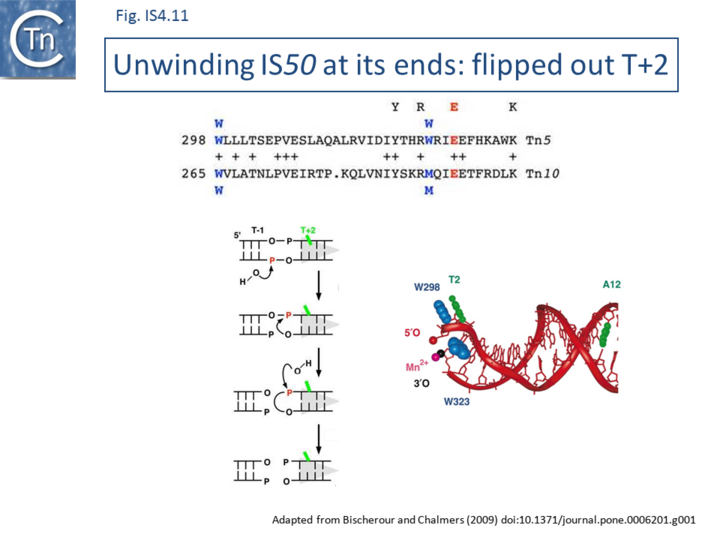
The Tn5 transpososome structure provided an understanding of the hairpin formation and resolution reactions. The structure revealed an interesting deformation in the DNA that included a flipped-out base at the second residue of the non-transferred strand. This provides a mechanism to reduce strain on the DNA in the short tight terminal hairpin bend and facilitates hairpin formation. The structure also provided information concerning individual transposase amino acids likely involved in assisting base extrusion or base flipping[4]. It involves two tyrosine residues capable of stacking with the flipped-out base[89][51][52] ((Fig.IS4.11). The IHF folded transpososome plays an important role in the hairpin resolution step[54].
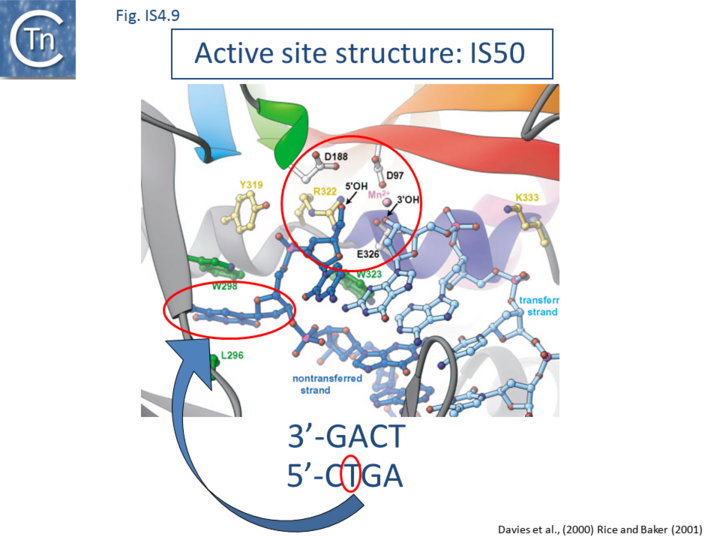
Base extrusion involves the YREK motif (Y-(2)-R-(3)-E-(6)-K) present in many DDE transposases and originally identified in the IS4 family[3]. The E of YREK is the catalytic E of the DDE motif. In the IS50 transposase structure, the Y, R, and K form contacts with the transposon end. Just after R in the YREK motif is a W residue (W323) which is inserted into the DNA minor groove. Hairpin formation during Tn5 transposition is assisted by two Trp residues acting in a “push-pull” mechanism (Bischerour and Chalmers, 2007). W323 may push the base to be flipped out whereas a second W, W298, captures this base, presumably by base stacking. In the Tn10 transposase W323 downstream of the R of the YREK motif, is substituted for an M motif that may have a similar role in extruding the flipped-out base[47][90][51].
Similar DNA hairpin stabilization by base flipping has been proposed for V(D)J recombination and transposition of Hermes 363 and Tn10[91][92].
IS231
The only other IS4 family member which has received some attention is IS231. This was isolated flanking a γ-endotoxin crystal protein gene from B. thuringiensis (see [93] and it has been proposed that it forms part of a composite transposon. IS231A is active in E. coli. Like IS10 and IS50, a potential ribosome binding site for the Tpase gene would be sequestered in a secondary structure in transcripts originating outside the element. Although most examples carry a single open reading frame, Tpase expression from two elements (IS231V and IS231W) may occur by a +1 and +2 frameshift respectively, but this has yet to be confirmed. Little is known about the transposition mechanism of this element although it exhibits strong target specificity with a preference for the ends of transposon Tn4430[94] and present evidence suggests that it transposes by a non-replicative cut and paste mechanism[95].
Several mobile cassettes derived from IS231 have been identified among Bacillus cereus and B. thuringiensis strains[96]. These elements consist of 50 to 80 bp, corresponding to the ends of various iso-IS231, flanking genes unrelated to transposition (e.g. adp, a D-stereospecific endopeptidase gene). At least in one case, such a cassette (known as MIC231) was shown to be trans-complemented by the transposase of IS231A[97].
IS701 family
The IS701 family was distinguished from the IS4 family by a highly conserved 4 bp target duplication, 5’ YTAR 3’. MCL analysis also indicated that the Tpases form a defined and separate group and alignments indicated the absence of Y in the Tpase YREK motif. There are several clades within this family. A new clade, ISAba11, was proposed as a new family based on 5 IS[98]. Members of this group generate 5 bp target duplications (instead of 4), exhibit conserved IR and include HHEK instead of YREK. However, additional examples exhibiting the conserved IR did not universally contain HHEK and MCL cluster analysis did not strongly support the notion that ISAba11 constitutes a new family. At present, we have retained ISAba11 as a subgroup in the IS701 family.
ISH3 family
The ISH3 family is restricted to the Archaea. In roughly half of the 30 members identified, the Tpase lacked the K/R residue of the DDE motif while all except ISFac10 displayed a Y(2)R(3)E(3)R motif. A characteristic of this family is the presence of 5 bp DR flanked at one end by A and at the other by T.
IS1634 family
The IS1634 family (previously IS1549) is characterized by large Tpases due to a β-strand insertion domain located between the conserved second D and E residues which is 35 to 79 aa longer than that of IS4 and members of the other related families. They generate 5-6 bp AT rich DR and are present in both archaea and bacteria[1]. Certain members generate very long variable DR (e.g. IS1634 from 17 to 478 bp[6]; ISCsa8 from 16 to 131 bp; ISMhp1, 80 bp).
Acknowledgements
We would like to thank Phoebe Rice (University of Chicago), David Haniford (University of Western Ontario), Ronald Chalmers (University of Nottingham) and Jacques Mahillon (Université de Lovain la Neuve) for permission to use figures in this chapter.
Bibliography
- ↑ 1.0 1.1 1.2 De Palmenaer D, Siguier P, Mahillon J . IS4 family goes genomic. - BMC Evol Biol: 2008 Jan 23, 8;18 [PubMed:18215304] [DOI]
- ↑ 2.0 2.1 Hickman AB, Chandler M, Dyda F . Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. - Crit Rev Biochem Mol Biol: 2010 Feb, 45(1);50-69 [PubMed:20067338] [DOI]
- ↑ 3.0 3.1 3.2 Rezsöhazy R, Hallet B, Delcour J, Mahillon J . The IS4 family of insertion sequences: evidence for a conserved transposase motif. - Mol Microbiol: 1993 Sep, 9(6);1283-95 [PubMed:7934941] [DOI]
- ↑ 4.0 4.1 4.2 4.3 Davies DR, Goryshin IY, Reznikoff WS, Rayment I . Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. - Science: 2000 Jul 7, 289(5476);77-85 [PubMed:10884228] [DOI]
- ↑ Mazel D, Bernard C, Schwarz R, Castets AM, Houmard J, Tandeau de Marsac N . Characterization of two insertion sequences, IS701 and IS702, from the cyanobacterium Calothrix species PCC 7601. - Mol Microbiol: 1991 Sep, 5(9);2165-70 [PubMed:1662761] [DOI]
- ↑ 6.0 6.1 Vilei EM, Nicolet J, Frey J . IS1634, a novel insertion element creating long, variable-length direct repeats which is specific for Mycoplasma mycoides subsp. mycoides small-colony type. - J Bacteriol: 1999 Feb, 181(4);1319-23 [PubMed:9973360] [DOI]
- ↑ Klenchin VA, Czyz A, Goryshin IY, Gradman R, Lovell S, Rayment I, Reznikoff WS . Phosphate coordination and movement of DNA in the Tn5 synaptic complex: role of the (R)YREK motif. - Nucleic Acids Res: 2008 Oct, 36(18);5855-62 [PubMed:18790806] [DOI]
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 Haniford DB, Ellis MJ . Transposons Tn10 and Tn5. - Microbiol Spectr: 2015 Feb, 3(1);MDNA3-0002-2014 [PubMed:26104553] [DOI]
- ↑ 9.0 9.1 Reznikoff WS . The Tn5 transposon. - Annu Rev Microbiol: 1993, 47;945-63 [PubMed:7504907] [DOI]
- ↑ Reznikoff WS, Bhasin A, Davies DR, Goryshin IY, Mahnke LA, Naumann T, Rayment I, Steiniger-White M, Twining SS . Tn5: A molecular window on transposition. - Biochem Biophys Res Commun: 1999 Dec 29, 266(3);729-34 [PubMed:10603311] [DOI]
- ↑ Reznikoff WS . Transposon Tn5. - Annu Rev Genet: 2008, 42;269-86 [PubMed:18680433] [DOI]
- ↑ Chalmers R, Sewitz S, Lipkow K, Crellin P . Complete nucleotide sequence of Tn10. - J Bacteriol: 2000 May, 182(10);2970-2 [PubMed:10781570] [DOI]
- ↑ 13.0 13.1 Rothstein SJ, Jorgensen RA, Postle K, Reznikoff WS . The inverted repeats of Tn5 are functionally different. - Cell: 1980 Mar, 19(3);795-805 [PubMed:6244898] [DOI]
- ↑ Rothstein SJ, Reznikoff WS . The functional differences in the inverted repeats of Tn5 are caused by a single base pair nonhomology. - Cell: 1981 Jan, 23(1);191-9 [PubMed:6260374] [DOI]
- ↑ Johnson RC, Yin JC, Reznikoff WS . Control of Tn5 transposition in Escherichia coli is mediated by protein from the right repeat. - Cell: 1982 Oct, 30(3);873-82 [PubMed:6291786] [DOI]
- ↑ Way JC, Davis MA, Morisato D, Roberts DE, Kleckner N . New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. - Gene: 1984 Dec, 32(3);369-79 [PubMed:6099322] [DOI]
- ↑ Halling SM, Simons RW, Way JC, Walsh RB, Kleckner N . DNA sequence organization of IS10-right of Tn10 and comparison with IS10-left. - Proc Natl Acad Sci U S A: 1982 Apr, 79(8);2608-12 [PubMed:6283536] [DOI]
- ↑ Johnson RC, Reznikoff WS . DNA sequences at the ends of transposon Tn5 required for transposition. - Nature: 1983 Jul 21-27, 304(5923);280-2 [PubMed:6306482] [DOI]
- ↑ Davis MA, Simons RW, Kleckner N . Tn10 protects itself at two levels from fortuitous activation by external promoters. - Cell: 1985 Nov, 43(1);379-87 [PubMed:2416461] [DOI]
- ↑ Schulz VP, Reznikoff WS . Translation initiation of IS50R read-through transcripts. - J Mol Biol: 1991 Sep 5, 221(1);65-80 [PubMed:1717696] [DOI]
- ↑ 21.0 21.1 21.2 Krebs MP, Reznikoff WS . Transcriptional and translational initiation sites of IS50. Control of transposase and inhibitor expression. - J Mol Biol: 1986 Dec 20, 192(4);781-91 [PubMed:2438419] [DOI]
- ↑ Yin JC, Krebs MP, Reznikoff WS . Effect of dam methylation on Tn5 transposition. - J Mol Biol: 1988 Jan 5, 199(1);35-45 [PubMed:2451025] [DOI]
- ↑ Kittle JD, Simons RW, Lee J, Kleckner N . Insertion sequence IS10 anti-sense pairing initiates by an interaction between the 5' end of the target RNA and a loop in the anti-sense RNA. - J Mol Biol: 1989 Dec 5, 210(3);561-72 [PubMed:2482367] [DOI]
- ↑ 24.0 24.1 Case CC, Roels SM, Jensen PD, Lee J, Kleckner N, Simons RW . The unusual stability of the IS10 anti-sense RNA is critical for its function and is determined by the structure of its stem-domain. - EMBO J: 1989 Dec 20, 8(13);4297-305 [PubMed:2480235] [DOI]
- ↑ Simons RW, Hoopes BC, McClure WR, Kleckner N . Three promoters near the termini of IS10: pIN, pOUT, and pIII. - Cell: 1983 Sep, 34(2);673-82 [PubMed:6311437] [DOI]
- ↑ Kleckner N, Way J, Davis M, Simons R, Halling S . Transposon Tn10: genetic organization, regulation, and insertion specificity. - Fed Proc: 1982 Aug, 41(10);2649-52 [PubMed:6286364]
- ↑ Tomizawa J . Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. - Cell: 1984 Oct, 38(3);861-70 [PubMed:6207934] [DOI]
- ↑ Tomizawa J, Som T . Control of ColE1 plasmid replication: enhancement of binding of RNA I to the primer transcript by the Rom protein. - Cell: 1984 Oct, 38(3);871-8 [PubMed:6207935] [DOI]
- ↑ Bushman FD, Ptashne M . Activation of transcription by the bacteriophage 434 repressor. - Proc Natl Acad Sci U S A: 1986 Dec, 83(24);9353-7 [PubMed:3467310] [DOI]
- ↑ Signon L, Kleckner N . Negative and positive regulation of Tn10/IS10-promoted recombination by IHF: two distinguishable processes inhibit transposition off of multicopy plasmid replicons and activate chromosomal events that favor evolution of new transposons. - Genes Dev: 1995 May 1, 9(9);1123-36 [PubMed:7744253] [DOI]
- ↑ 31.0 31.1 31.2 Sakai JS, Kleckner N, Yang X, Guhathakurta A . Tn10 transpososome assembly involves a folded intermediate that must be unfolded for target capture and strand transfer. - EMBO J: 2000 Feb 15, 19(4);776-85 [PubMed:10675347] [DOI]
- ↑ 32.0 32.1 32.2 32.3 Chalmers R, Guhathakurta A, Benjamin H, Kleckner N . IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. - Cell: 1998 May 29, 93(5);897-908 [PubMed:9630232] [DOI]
- ↑ 33.0 33.1 Swingle B, O'Carroll M, Haniford D, Derbyshire KM . The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10. - Mol Microbiol: 2004 May, 52(4);1055-67 [PubMed:15130124] [DOI]
- ↑ Navarre WW, McClelland M, Libby SJ, Fang FC . Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. - Genes Dev: 2007 Jun 15, 21(12);1456-71 [PubMed:17575047] [DOI]
- ↑ Higashi K, Tobe T, Kanai A, Uyar E, Ishikawa S, Suzuki Y, Ogasawara N, Kurokawa K, Oshima T . H-NS Facilitates Sequence Diversification of Horizontally Transferred DNAs during Their Integration in Host Chromosomes. - PLoS Genet: 2016 Jan, 12(1);e1005796 [PubMed:26789284] [DOI]
- ↑ 36.0 36.1 36.2 36.3 Wardle SJ, Chan A, Haniford DB . H-NS binds with high affinity to the Tn10 transpososome and promotes transpososome stabilization. - Nucleic Acids Res: 2009 Oct, 37(18);6148-60 [PubMed:19696075] [DOI]
- ↑ 37.0 37.1 37.2 Wardle SJ, O'Carroll M, Derbyshire KM, Haniford DB . The global regulator H-NS acts directly on the transpososome to promote Tn10 transposition. - Genes Dev: 2005 Sep 15, 19(18);2224-35 [PubMed:16166383] [DOI]
- ↑ 38.0 38.1 38.2 Ward CM, Wardle SJ, Singh RK, Haniford DB . The global regulator H-NS binds to two distinct classes of sites within the Tn10 transpososome to promote transposition. - Mol Microbiol: 2007 May, 64(4);1000-13 [PubMed:17501923] [DOI]
- ↑ 39.0 39.1 39.2 39.3 Whitfield CR, Wardle SJ, Haniford DB . The global bacterial regulator H-NS promotes transpososome formation and transposition in the Tn5 system. - Nucleic Acids Res: 2009 Feb, 37(2);309-21 [PubMed:19042975] [DOI]
- ↑ 40.0 40.1 Liu D, Haniford DB, Chalmers RM . H-NS mediates the dissociation of a refractory protein-DNA complex during Tn10/IS10 transposition. - Nucleic Acids Res: 2011 Aug, 39(15);6660-8 [PubMed:21565798] [DOI]
- ↑ 41.0 41.1 Singh RK, Liburd J, Wardle SJ, Haniford DB . The nucleoid binding protein H-NS acts as an anti-channeling factor to favor intermolecular Tn10 transposition and dissemination. - J Mol Biol: 2008 Feb 29, 376(4);950-62 [PubMed:18191147] [DOI]
- ↑ 42.0 42.1 Mizuuchi M, Rice PA, Wardle SJ, Haniford DB, Mizuuchi K . Control of transposase activity within a transpososome by the configuration of the flanking DNA segment of the transposon. - Proc Natl Acad Sci U S A: 2007 Sep 11, 104(37);14622-7 [PubMed:17785414] [DOI]
- ↑ 43.0 43.1 Humayun S, Wardle SJ, Shilton BH, Pribil PA, Liburd J, Haniford DB . Tn10 transposase mutants with altered transpososome unfolding properties are defective in hairpin formation. - J Mol Biol: 2005 Feb 25, 346(3);703-16 [PubMed:15713457] [DOI]
- ↑ 44.0 44.1 Pribil PA, Wardle SJ, Haniford DB . Enhancement and rescue of target capture in Tn10 transposition by site-specific modifications in target DNA. - Mol Microbiol: 2004 May, 52(4);1173-86 [PubMed:15130133] [DOI]
- ↑ 45.0 45.1 Pribil PA, Haniford DB . Target DNA bending is an important specificity determinant in target site selection in Tn10 transposition. - J Mol Biol: 2003 Jul 4, 330(2);247-59 [PubMed:12823965] [DOI]
- ↑ Stewart BJ, Wardle SJ, Haniford DB . IHF-independent assembly of the Tn10 strand transfer transpososome: implications for inhibition of disintegration. - EMBO J: 2002 Aug 15, 21(16);4380-90 [PubMed:12169640] [DOI]
- ↑ 47.0 47.1 Allingham JS, Wardle SJ, Haniford DB . Determinants for hairpin formation in Tn10 transposition. - EMBO J: 2001 Jun 1, 20(11);2931-42 [PubMed:11387226] [DOI]
- ↑ Pribil PA, Haniford DB . Substrate recognition and induced DNA deformation by transposase at the target-capture stage of Tn10 transposition. - J Mol Biol: 2000 Oct 20, 303(2);145-59 [PubMed:11023782] [DOI]
- ↑ Liu D, Chalmers R . Hyperactive mariner transposons are created by mutations that disrupt allosterism and increase the rate of transposon end synapsis. - Nucleic Acids Res: 2014 Feb, 42(4);2637-45 [PubMed:24319144] [DOI]
- ↑ Bischerour J, Chalmers R . Base flipping in tn10 transposition: an active flip and capture mechanism. - PLoS One: 2009 Jul 10, 4(7);e6201 [PubMed:19593448] [DOI]
- ↑ 51.0 51.1 51.2 51.3 Bischerour J, Chalmers R . Base-flipping dynamics in a DNA hairpin processing reaction. - Nucleic Acids Res: 2007, 35(8);2584-95 [PubMed:17412704] [DOI]
- ↑ 52.0 52.1 52.2 Liu D, Sewitz S, Crellin P, Chalmers R . Functional coupling between the two active sites during Tn 10 transposition buffers the mutation of sequences critical for DNA hairpin processing. - Mol Microbiol: 2006 Dec, 62(6);1522-33 [PubMed:17083470] [DOI]
- ↑ 53.0 53.1 Liu D, Crellin P, Chalmers R . Cyclic changes in the affinity of protein-DNA interactions drive the progression and regulate the outcome of the Tn10 transposition reaction. - Nucleic Acids Res: 2005, 33(6);1982-92 [PubMed:15814815] [DOI]
- ↑ 54.0 54.1 54.2 Crellin P, Sewitz S, Chalmers R . DNA looping and catalysis; the IHF-folded arm of Tn10 promotes conformational changes and hairpin resolution. - Mol Cell: 2004 Feb 27, 13(4);537-47 [PubMed:14992723] [DOI]
- ↑ 55.0 55.1 Sewitz S, Crellin P, Chalmers R . The positive and negative regulation of Tn10 transposition by IHF is mediated by structurally asymmetric transposon arms. - Nucleic Acids Res: 2003 Oct 15, 31(20);5868-76 [PubMed:14530435] [DOI]
- ↑ 56.0 56.1 Crellin P, Chalmers R . Protein-DNA contacts and conformational changes in the Tn10 transpososome during assembly and activation for cleavage. - EMBO J: 2001 Jul 16, 20(14);3882-91 [PubMed:11447129] [DOI]
- ↑ Lease RA, Belfort M . A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. - Proc Natl Acad Sci U S A: 2000 Aug 29, 97(18);9919-24 [PubMed:10954740] [DOI]
- ↑ 58.0 58.1 58.2 Ross JA, Wardle SJ, Haniford DB . Tn10/IS10 transposition is downregulated at the level of transposase expression by the RNA-binding protein Hfq. - Mol Microbiol: 2010 Nov, 78(3);607-21 [PubMed:20815820] [DOI]
- ↑ Ellis MJ, Trussler RS, Haniford DB . Hfq binds directly to the ribosome-binding site of IS10 transposase mRNA to inhibit translation. - Mol Microbiol: 2015 May, 96(3);633-50 [PubMed:25649688] [DOI]
- ↑ 60.0 60.1 Ross JA, Ellis MJ, Hossain S, Haniford DB . Hfq restructures RNA-IN and RNA-OUT and facilitates antisense pairing in the Tn10/IS10 system. - RNA: 2013 May, 19(5);670-84 [PubMed:23510801] [DOI]
- ↑ Ellis MJ, Trussler RS, Ross JA, Haniford DB . Probing Hfq:RNA interactions with hydroxyl radical and RNase footprinting. - Methods Mol Biol: 2015, 1259;403-15 [PubMed:25579599] [DOI]
- ↑ Weinreich MD, Reznikoff WS . Fis plays a role in Tn5 and IS50 transposition. - J Bacteriol: 1992 Jul, 174(14);4530-7 [PubMed:1320613] [DOI]
- ↑ Mahnke Braam LA, Reznikoff WS . Functional characterization of the Tn5 transposase by limited proteolysis. - J Biol Chem: 1998 May 1, 273(18);10908-13 [PubMed:9556567] [DOI]
- ↑ de la Cruz NB, Weinreich MD, Wiegand TW, Krebs MP, Reznikoff WS . Characterization of the Tn5 transposase and inhibitor proteins: a model for the inhibition of transposition. - J Bacteriol: 1993 Nov, 175(21);6932-8 [PubMed:8226636] [DOI]
- ↑ 65.0 65.1 Haniford DB, Chelouche AR, Kleckner N . A specific class of IS10 transposase mutants are blocked for target site interactions and promote formation of an excised transposon fragment. - Cell: 1989 Oct 20, 59(2);385-94 [PubMed:2553270] [DOI]
- ↑ 66.0 66.1 66.2 Kwon D, Chalmers RM, Kleckner N . Structural domains of IS10 transposase and reconstitution of transposition activity from proteolytic fragments lacking an interdomain linker. - Proc Natl Acad Sci U S A: 1995 Aug 29, 92(18);8234-8 [PubMed:7667274] [DOI]
- ↑ Weinreich MD, Mahnke-Braam L, Reznikoff WS . A functional analysis of the Tn5 transposase. Identification of domains required for DNA binding and multimerization. - J Mol Biol: 1994 Aug 12, 241(2);166-77 [PubMed:8057357] [DOI]
- ↑ 68.0 68.1 Morisato D, Kleckner N . Tn10 transposition and circle formation in vitro. - Cell: 1987 Oct 9, 51(1);101-11 [PubMed:2820584] [DOI]
- ↑ 69.0 69.1 Chalmers RM, Kleckner N . Tn10/IS10 transposase purification, activation, and in vitro reaction. - J Biol Chem: 1994 Mar 18, 269(11);8029-35 [PubMed:8132525]
- ↑ 70.0 70.1 Goryshin IY, Reznikoff WS . Tn5 in vitro transposition. - J Biol Chem: 1998 Mar 27, 273(13);7367-74 [PubMed:9516433] [DOI]
- ↑ Bender J, Kleckner N . Genetic evidence that Tn10 transposes by a nonreplicative mechanism. - Cell: 1986 Jun 20, 45(6);801-15 [PubMed:3011280] [DOI]
- ↑ Benjamin HW, Kleckner N . Excision of Tn10 from the donor site during transposition occurs by flush double-strand cleavages at the transposon termini. - Proc Natl Acad Sci U S A: 1992 May 15, 89(10);4648-52 [PubMed:1316613] [DOI]
- ↑ Morisato D, Kleckner N . Transposase promotes double strand breaks and single strand joints at Tn10 termini in vivo. - Cell: 1984 Nov, 39(1);181-90 [PubMed:6091910] [DOI]
- ↑ Bolland S, Kleckner N . The two single-strand cleavages at each end of Tn10 occur in a specific order during transposition. - Proc Natl Acad Sci U S A: 1995 Aug 15, 92(17);7814-8 [PubMed:7644497] [DOI]
- ↑ Bolland S, Kleckner N . The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. - Cell: 1996 Jan 26, 84(2);223-33 [PubMed:8565068] [DOI]
- ↑ 76.0 76.1 Bhasin A, Goryshin IY, Reznikoff WS . Hairpin formation in Tn5 transposition. - J Biol Chem: 1999 Dec 24, 274(52);37021-9 [PubMed:10601258] [DOI]
- ↑ 77.0 77.1 Kennedy AK, Guhathakurta A, Kleckner N, Haniford DB . Tn10 transposition via a DNA hairpin intermediate. - Cell: 1998 Oct 2, 95(1);125-34 [PubMed:9778253] [DOI]
- ↑ Kennedy AK, Haniford DB, Mizuuchi K . Single active site catalysis of the successive phosphoryl transfer steps by DNA transposases: insights from phosphorothioate stereoselectivity. - Cell: 2000 Apr 28, 101(3);295-305 [PubMed:10847684] [DOI]
- ↑ Bischerour J, Chalmers R . Base flipping in tn10 transposition: an active flip and capture mechanism. - PLoS One: 2009 Jul 10, 4(7);e6201 [PubMed:19593448] [DOI]
- ↑ Haniford DB . Transpososome dynamics and regulation in Tn10 transposition. - Crit Rev Biochem Mol Biol: 2006 Nov-Dec, 41(6);407-24 [PubMed:17092825] [DOI]
- ↑ Whitfield CR, Wardle SJ, Haniford DB . Formation, characterization and partial purification of a Tn5 strand transfer complex. - J Mol Biol: 2006 Dec 1, 364(3);290-301 [PubMed:17014865] [DOI]
- ↑ Dorman CJ . H-NS: a universal regulator for a dynamic genome. - Nat Rev Microbiol: 2004 May, 2(5);391-400 [PubMed:15100692] [DOI]
- ↑ Davies DR, Mahnke Braam L, Reznikoff WS, Rayment I . The three-dimensional structure of a Tn5 transposase-related protein determined to 2.9-A resolution. - J Biol Chem: 1999 Apr 23, 274(17);11904-13 [PubMed:10207011] [DOI]
- ↑ Naumann TA, Reznikoff WS . Trans catalysis in Tn5 transposition. - Proc Natl Acad Sci U S A: 2000 Aug 1, 97(16);8944-9 [PubMed:10908658] [DOI]
- ↑ 85.0 85.1 85.2 Mahnke Braam LA, Goryshin IY, Reznikoff WS . A mechanism for Tn5 inhibition. carboxyl-terminal dimerization. - J Biol Chem: 1999 Jan 1, 274(1);86-92 [PubMed:9867814] [DOI]
- ↑ York D, Reznikoff WS . Purification and biochemical analyses of a monomeric form of Tn5 transposase. - Nucleic Acids Res: 1996 Oct 1, 24(19);3790-6 [PubMed:8871560] [DOI]
- ↑ Duval-Valentin G, Chandler M . Cotranslational control of DNA transposition: a window of opportunity. - Mol Cell: 2011 Dec 23, 44(6);989-96 [PubMed:22195971] [DOI]
- ↑ Morisato D, Way JC, Kim HJ, Kleckner N . Tn10 transposase acts preferentially on nearby transposon ends in vivo. - Cell: 1983 Mar, 32(3);799-807 [PubMed:6299577] [DOI]
- ↑
- ↑ Bischerour J, Chalmers R . Base flipping in tn10 transposition: an active flip and capture mechanism. - PLoS One: 2009 Jul 10, 4(7);e6201 [PubMed:19593448] [DOI]
- ↑ Lu CP, Sandoval H, Brandt VL, Rice PA, Roth DB . Amino acid residues in Rag1 crucial for DNA hairpin formation. - Nat Struct Mol Biol: 2006 Nov, 13(11);1010-5 [PubMed:17028591] [DOI]
- ↑ Mahillon J, Rezsöhazy R, Hallet B, Delcour J . IS231 and other Bacillus thuringiensis transposable elements: a review. - Genetica: 1994, 93(1-3);13-26 [PubMed:7813910] [DOI]
- ↑ Hallet B, Rezsöhazy R, Delcour J . IS231A from Bacillus thuringiensis is functional in Escherichia coli: transposition and insertion specificity. - J Bacteriol: 1991 Jul, 173(14);4526-9 [PubMed:1648561] [DOI]
- ↑ Léonard C, Mahillon J . IS231A transposition: conservative versus replicative pathway. - Res Microbiol: 1998 Sep, 149(8);549-55 [PubMed:9795992] [DOI]
- ↑ De Palmenaer D, Vermeiren C, Mahillon J . IS231-MIC231 elements from Bacillus cereus sensu lato are modular. - Mol Microbiol: 2004 Jul, 53(2);457-67 [PubMed:15228527] [DOI]
- ↑ Chen Y, Braathen P, Léonard C, Mahillon J . MIC231, a naturally occurring mobile insertion cassette from Bacillus cereus. - Mol Microbiol: 1999 May, 32(3);657-68 [PubMed:10320586] [DOI]
- ↑ Rieck B, Tourigny DS, Crosatti M, Schmid R, Kochar M, Harrison EM, Ou HY, Turton JF, Rajakumar K . Acinetobacter insertion sequence ISAba11 belongs to a novel family that encodes transposases with a signature HHEK motif. - Appl Environ Microbiol: 2012 Jan, 78(2);471-80 [PubMed:22081580] [DOI]









