IS Families/IS1202 family
This chapter has appeared in a modified form as: A subclass of the IS1202 family of bacterial insertion sequences targets XerCD recombination sites. Patricia Siguier, Philippe Rousseau, François Cornet, Michael Chandler. Plasmid 2023 Jun 9;127:102696. doi:10.1016/j.plasmid.2023.102696.[1]
Contents
The IS1202 Family
History
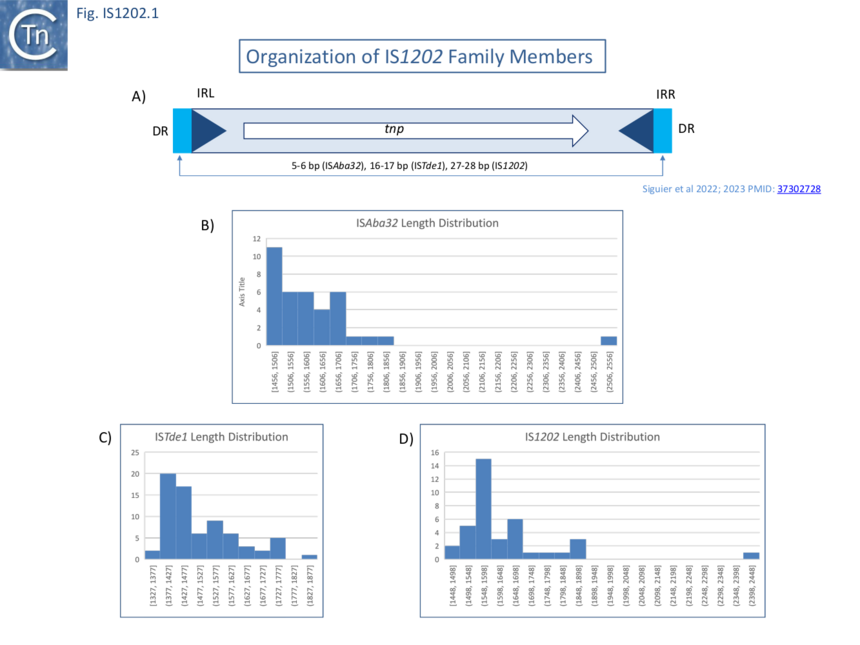
The founding member of this family, the 1,747 bp IS1202, identified in Streptococcus pneumoniae [2], is bordered by 23 bp imperfect inverted repeat sequences (IR), contains a single open reading frame sufficient to encode a 54.4-kDa polypeptide and is flanked by a 27 bp direct target repeat sequence (DR). IS1202 was initially classified in ISfinder as an emerging IS family (ISNCY – not classified yet) [3] (See IS families table at: TnPedia) but further genome analysis [4][1][5] identified over 150 related examples which, together, constitute the IS1202 family.
Many were identified in public databases by reiterative BLAST approaches [6] with the primary transposase sequence of representative elements used as a query in a BLASTP [7]. The collection of IS forms a coherent family, the IS1202 family, based on their transposase sequences [4][1]. While certain have impacted some important properties of their hosts (e.g. deletion of the capsular polysaccharide locus (cps), a major Streptococcus pneumoniae virulence factor, key to survival in the blood [8] and insertional inactivation of lipopolysaccharide gene, lpxA, resulting in colistin dependence possibly leading to colistin resistance [9]), a potentially more interesting property is that a specific subclass of the family targets xrs (xer) recombination sites (Target-specificity of ISAba32 subgroup members), including the universal chromosome termination site, dif (see for example [10]).
Organization, Phylogenetic Analysis and Identification of Three Major Subgroups.
Family members range from 1,320 to 1,990 bp in length with a single Tpase orf of between 400 and 500 amino acids long in a single reading frame (Fig. IS1202.1 A-D). Several, (i.e. tISKpn21, tISKpn65, tISKpn63, tISShal2 and tISRel10) also carry a single passenger gene, annotated as “hypothetical protein”, which is unrelated in each case.
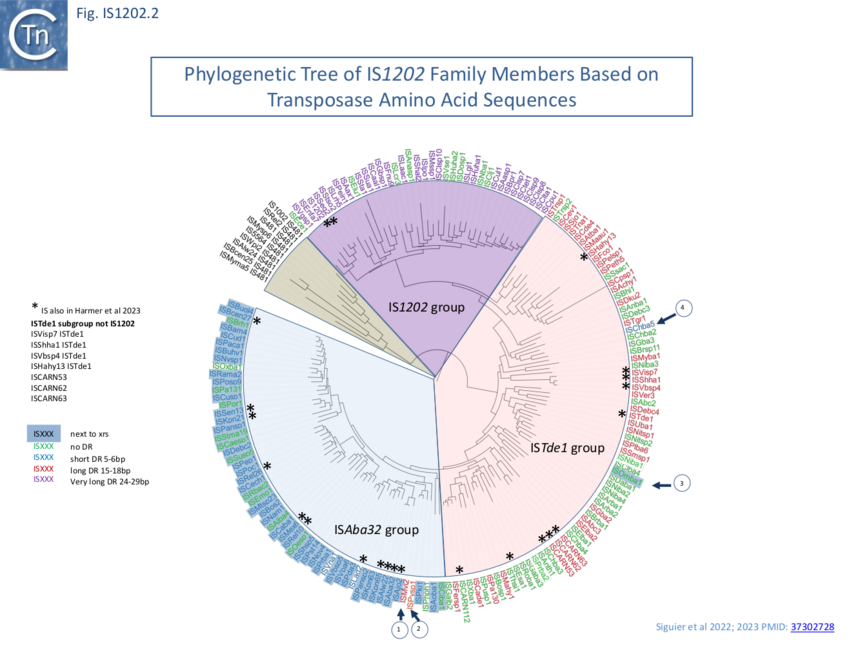
The examples fall into three principal subgroups defined by Tpase alignments (Fig. IS1202.2). Each has been named after one of their members as: IS1202, ISTde1 and ISAba32 [4][1]. Note that these groups are largely similar to those proposed by Harmer et al [5] with slight differences presumably because those shown here are based on a larger IS library: the ISAba32 (61 examples;[4][1]) and ISAjo2 (16 examples; [5]) groups are equivalent, whereas ISCARN52, ISCARN62 and ISCARN63 and several others which appear in the IS1202 subgroup of Harmer et al [5] (10 examples) do not fall into the IS1202 subgroup of Siguier et al [4][1] (38 examples;), rather they are included in the ISTde1 subgroup along with ISCARN112 and ISEsa1 (ISEsa1 subgroup; 2 examples; [5]). A phylogenetic tree, rooted with members of the IS481 family is shown in Fig. IS1202.2. The four family members which also carry apparently unrelated passenger genes are not restricted to a single subgroup: for example, whereas tISKpn21, tISKpn65, tISKpn63, and tISRel10 all belong to the ISAba32 subgroup, tISShal2 belongs to the IS1202 subgroup.
Direct Target Repeat Length
Members of the three IS1202 family subgroups also generate DRs of three different lengths: Short, 5-6 bp (ISAba32); Medium, 15-18 bp (ISTde1); and Long 24-29 bp, (IS1202) (Fig. IS1202.1 and Fig.IS1202.2). In the case of a few examples, no direct repeats are present. However, in many cases, other copies of the same IS identified elsewhere did exhibit DRs [4][1]. The absence of DR in these cases could therefore simply be the result of intra-replicon recombination between two resident IS copies, leading to the separation of the flanking DR sequences, the result from genetic drift in the DR or, since all were found in genomes assembled from shotgun sequencing, by errors of assembly. Similar results were obtained by Harmer et al [5].
Transposase Signatures
A domain search (COG and HMMER/PFAM and a de novo search with MEME) revealed two major domains: an N-terminal helix-turn-helix (HTH) DNA-binding domain and a DDE-type RNase fold catalytic domain (Fig. IS1202. 3A).
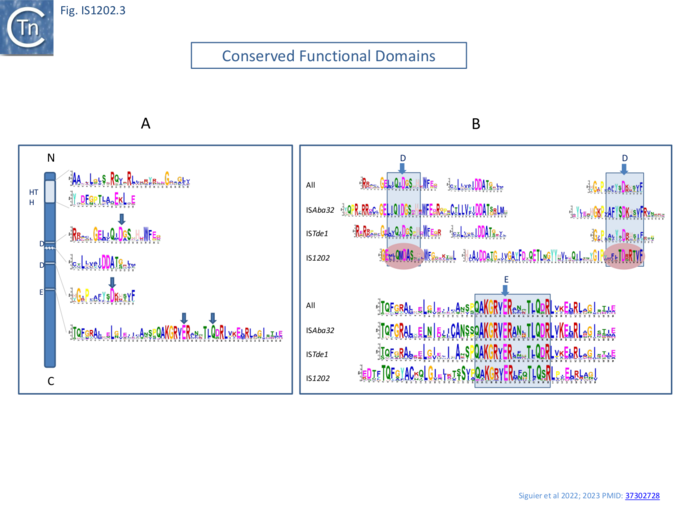
Within the DDE domain, there are two highly conserved additional Aspartic acid (DD) residues between the two D and a glutamine (Q) seven residues C-terminal to the conserved E (Glutamic acid) instead of the characteristic K/R (Lysine/Arginine) [3][11] as also noted by Harmer et al [5]. The motifs surrounding the DDE triad are retained by each of the subgroups individually (Fig.IS1202.3B).
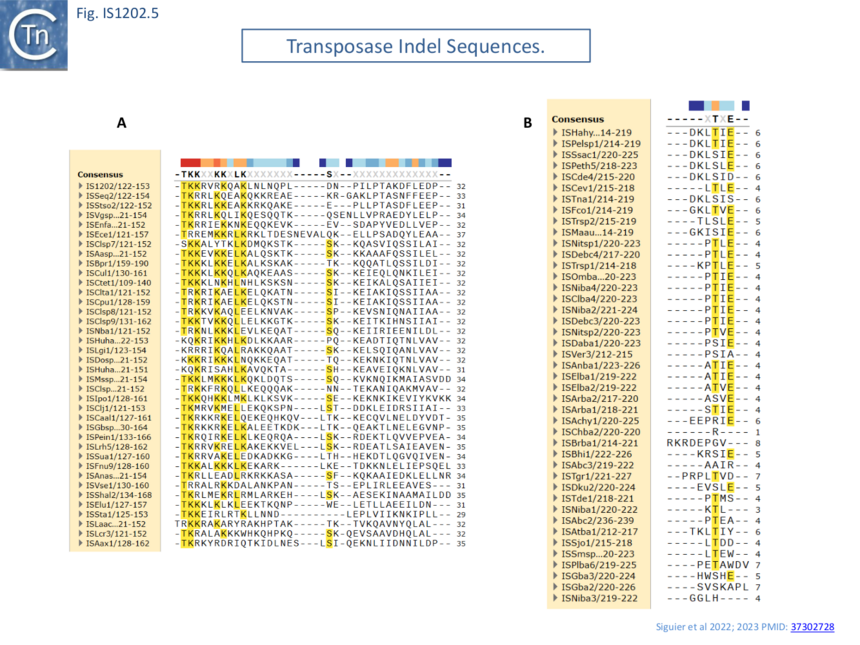
Transposase alignment revealed two prominent group-specific indel sequences (Fig. IS1202.4): one of about 30 amino acids just before the catalytic domain (IS1202 subgroup), and a second smaller indel sequence of 1-7 amino acids (ISTde1 subgroup) between the second D and the E of the DDE domain. There was significant amino acid conservation in the larger indel sequence particularly at the N-terminal end (Fig. IS1202.5). All three subgroups also included a non-conserved C-terminal region (Fig. IS1202.4).
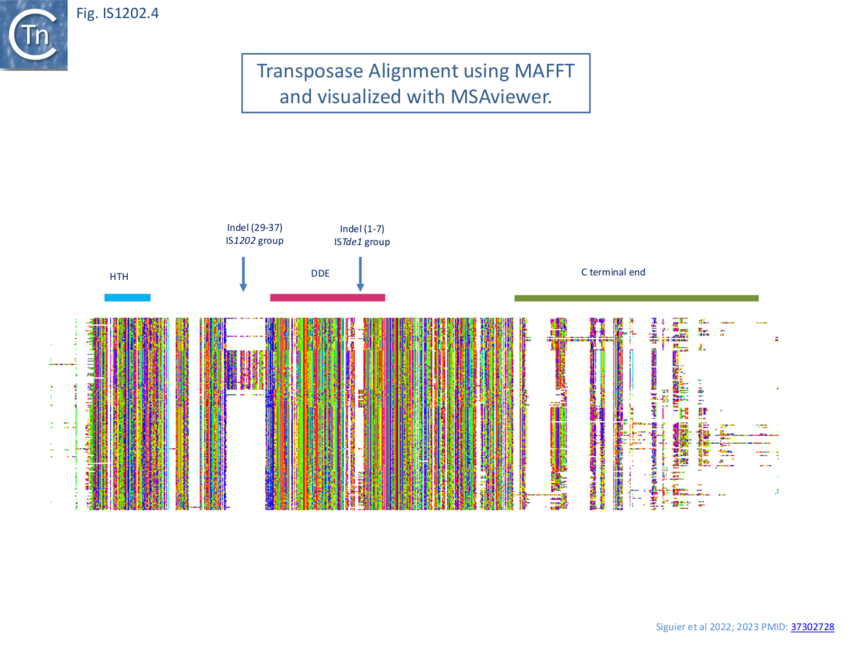
AlphaFold [12] models (Fig. IS1202.6) revealed that the N-terminal HTH (Helix-Turn-Helix) domain, presumably involved in IR binding, is separated from the catalytic domain carrying the catalytic site by a poorly defined segment (blue arrows). The variable C-terminal segment, predicted to be α-helical is also poorly defined (orange arrows) as a region of low or very low predictive confidence. For ISTde1, the insertion splits the DDE motif. In all three cases, the N-terminal HTH domain appears separated from the rest of the transposase by a region of low predictive confidence. These indels are correlated with mechanistic changes in transposition strictly associated with the behavior of the IS1202 subgroup in which they are found viz: change in xrs targeting and of the associated DR length.
The transposases of the IS1202 family appear related, although distantly, to that of the to IS481 and IS3 family particularly in their DDE domains (e.g. IS1202 transposase has 39% amino acid similarity to those of ISPfr5 of the IS481 family) [11].
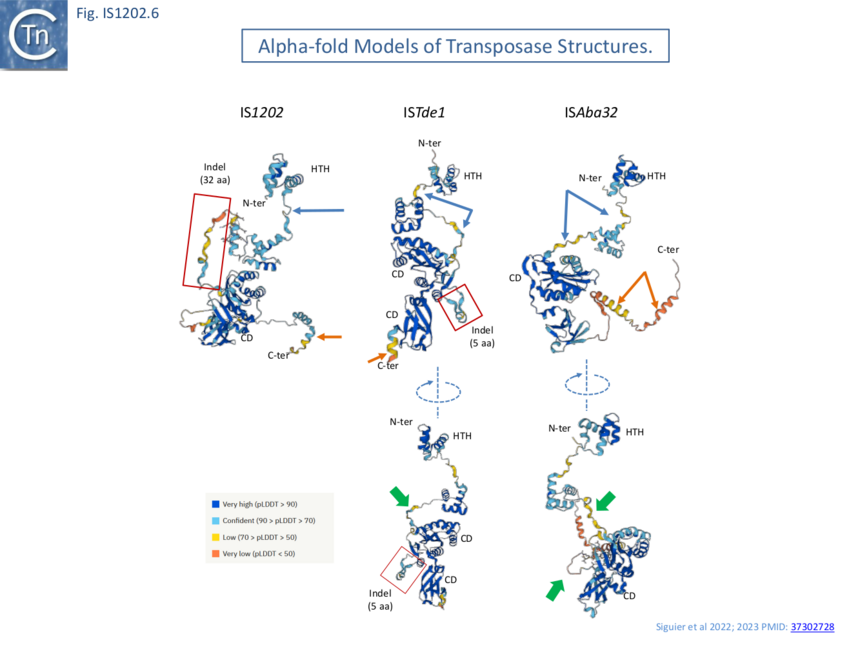
Harmer et al [5] suggest that IS1202 family transposases show some similarity to the phage Mu and Tn7 transposases although the resemblance of the DDE domain at the sequence level is not particularly strong (Fig. IS1202.7 top). However, HHpred analysis (Fig. IS1202. 7) indeed shows good structural similarities with Tn7 TnsB and MuA proteins as well as to the transposase of the eukaryotic mariner element Mos1. Harmer et al [5] have also underlined the presence of two N-Ter HTH modules composing the probable DNA binding domain. These can be seen in the AlphaFold models of Fig. IS1202.6. (ISAba32: https://alphafold.ebi.ac.uk/entry/A0A5N5XUG9; ISTde1:https://alphafold.ebi.ac.uk/entry/Q73JR2; IS1202: https://alphafold.ebi.ac.uk/entry/Q54513).
It should be noted that a number of transposases exhibit tandem DNA binding domains: Mos1 also includes 2 N-terminal HTH modules [13] as does the transposase of IS21 (Fig. IS21.7) [14] whereas Tn7 TnsB carries N-Ter SH3 (beta-barrel) and HTH modules with a winged helix DBD further downstream (Fig. Tn7.2Fi).

Terminal Inverted IRs
Alignment of the left and right terminal inverted repeats of each IS subgroup (Fig. IS1202.8) shows that, like the IRs of most IS (see: TnPedia), IS1202 family IRs carry two well conserved domains: a terminal domain of three base pairs, which is recognized for cleavage, and an internal region which generally serves as a DNA recognition sequence for transposase binding. The terminal domain of both IRL and IRR of members of two subgroups (ISAba32 and ISTde1) begins with 5’-TGT-3’ (as do those of the IS3 and IS481 families [11]) while those of the third subgroup, IS1202, are less conserved: IRR retains the conserved TGT, but the left end is less conserved (5’-Ta/gT-3’) (Fig. IS1202.9 A, B and C).
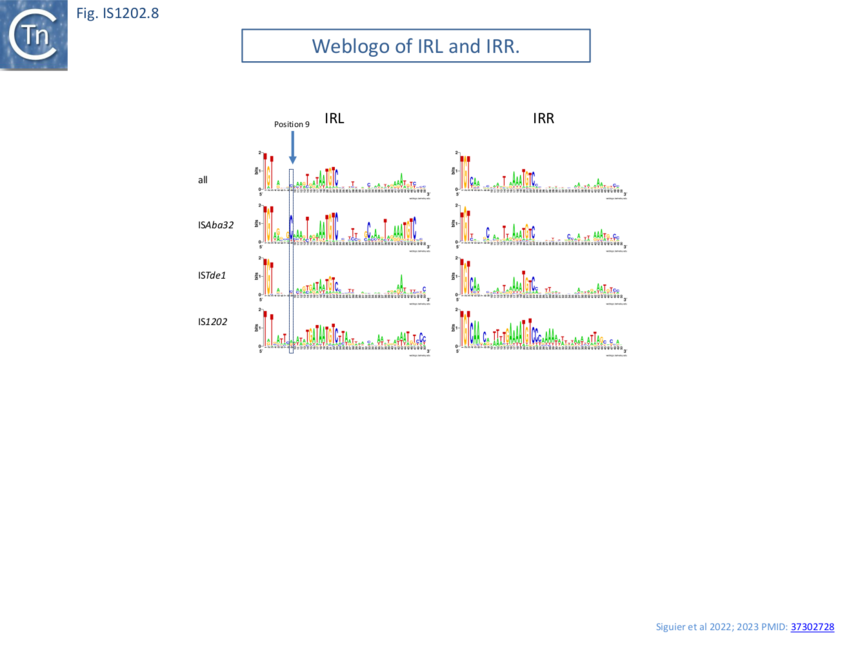
All three subgroups have a second conserved region around position 20 of both IRL and IRR. This is somewhat more extensive for the IS1202 subgroup than for the other two subgroups. Not only does the ISAba32 subgroup carry a third relatively well conserved region further into the IR, but it exhibits a completely conserved C residue at position 9 of IRL. Close examination of the end sequences (Fig. IS1202.9 A, B and C) revealed that the two conserved regions of the ISAba32 group represent two tandem and partially conserved direct repeats (also noted in reference [5]) which, in other transposons and IS, constitute transposase binding sites (e.g Tn7: Tn7.1A and the related Tn402: Fig.Tn402.1; Tn7: Fig.Tn7.1D; IS21: Fig. IS21.3; Fig. IS21.7B). As shown in Fig. IS1202.8, these sequences are less conserved in the right end than in IRL. There is a “core” conserved heptanucleotide block 5’-AAATGTC-3’ with some variation in the initial 3 nucleotides. The IS1202 subgroup has two copies of the core sequence in IRL but only a single copy in IRR (Fig. IS1202.8; Fig. IS1202.9 D). In this case the copy proximal to the IRL tip is frequently 5’-TAATGTC-3’ and can be extended slightly in both 5’ and 3’ directions. The third subgroup, ISTde1, has a slightly longer core repeat of 9 bp which, in many cases can be extended by a single base pair at the 5’ and 3’ ends (Fig. IS1202.9 F and F).
Thus, in addition to DR length and differences in the transposase, the IR of each IS1202 subgroup have distinct features.
Distribution.
The distribution of members of the three subgroups is quite different [4][1]: ISAba32 subgroup members are found in plasmids and chromosomes and in unassembled shotgun sequences of mainly γ- (Acinetobacter), β- (Burkholderia) and some α-proteobacteria; the majority of ISTde1 subgroup members were identified in whole shotgun sequences, in a number of chromosomes but in only one plasmid. They are more widely dispersed and can be found in α and β-proteobacteria, Firmicutes, Armatimonadota, Acidobacteria, Atribacterota, Chloroflexota, Deltaproteobacteria, Elusimicrobiota, Gemmatimonadetes, Nitrospira and Synergistota; the IS1202 subgroup is found in whole shotgun sequences and also in assembled chromosomes. Members have yet been identified in plasmids. They are found in Firmicutes, Tenericutes and a Spirochaete [4][1].
Target-specificity of ISAba32 subgroup members
For each IS, at least one (and frequently several) insertion sites corresponding to insertions in different loci were annotated generating a library of 245 insertions with their flanking sequences [4][1].
Xrs sites
Xrs (Xer Recombination Sites) is the generic name for specific recombination sites found on chromosomes and plasmids, acted on by the XerC and XerD recombinases [10]. XerCD recombine chromosome- and plasmid-borne xrs to resolve dimers formed by recombination between circular sister replicons (commonly known as dif sites for chromosomes and xer sites for plasmids).
Other xrs are used to integrate bacteriophages or genomic islands into chromosomes. More recently numerous xrs have been found flanking mobile genes in plasmids, thus inferred to be involved in their mobility [15][16][17][18]. Mobile Genetic Elements inserted next to xrs have been repeatedly identified in plasmids of Acinetobacter baumannii often carrying repeated xrs (called pdif in these cases, because of their homology to the chromosomal xrs, dif) arranged in modules in which they flank one or a small number of genes, often including different clinically important carbapenemase-encoding bla-OXA genes [15][16][17][18]. Similar structures have been identified in a number of additional bacterial genera and species [19][20].
Insertions abutting xrs
A number of IS1202-related IS had been identified abutting xrs in bacterial plasmids and chromosomes. Initially, these were observed in plasmids of Acinetobacter baumannii [16][17][21] but now include many bacterial genera and species such Klebsiella pneumoniae and Burkholderia cenocepacia [4][1].
Only members of the ISAba32 subgroup target xrs
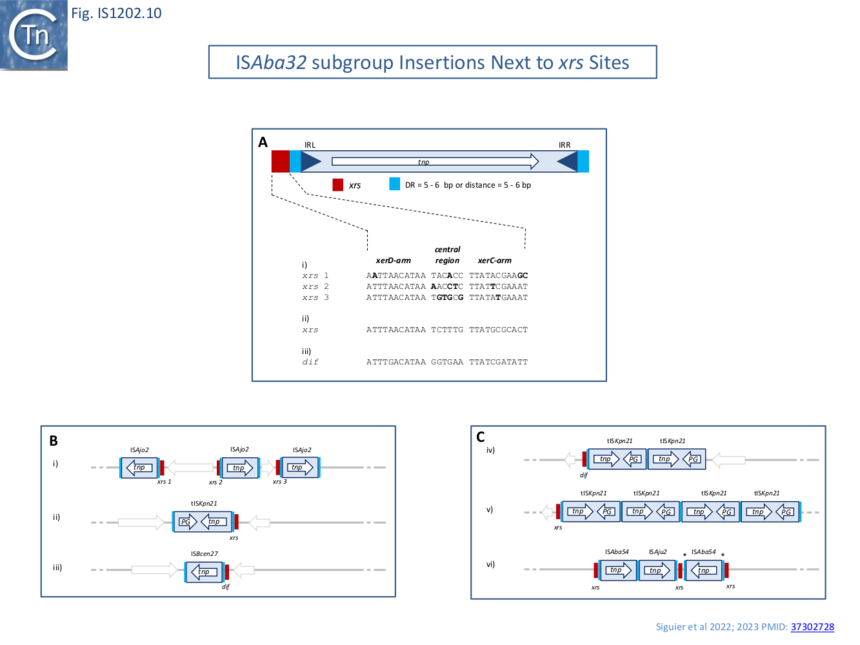
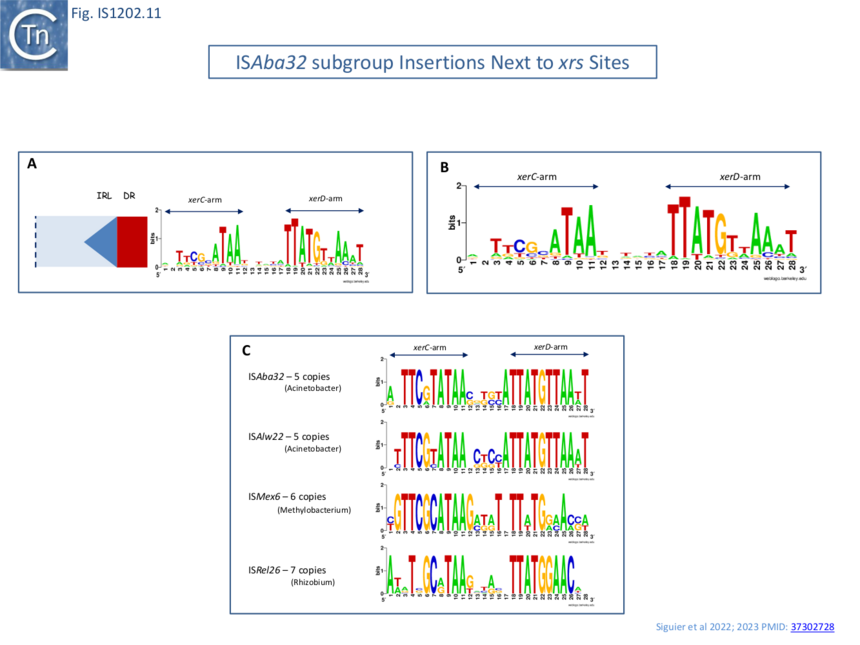
A large number of IS insertions neighboring xrs sites have now been identified [4][1][5] (Fig.IS1202.2, Fig.IS1202.10). They are invariably located 3-7 bp from the outer end of the XerC-arm of xrs, with the length corresponding to the DR (Fig.IS1202.10) and oriented with IRL next to the predicted XerC-binding arm of the xrs (Fig.IS1202.10, Fig.IS1202.11 A).
Only members of the ISAba32 subgroup have been observed inserted next to xrs (Fig. IS1202.2, [4][1]). In one study [1] 61 of 166 IS1202 family members, all belonging to the ISAba32 subgroup had inserted at 5-6 bp from an xrs (Fig. IS1202.10 Bii; Fig. IS1202.10 A). In most cases, a DR of 5-6 bp was detected. The insertions included both full length and partial ISAba32 copies and in each partial copy, IRL (but not IRR) was conserved and the distance between the xrs and the partial IS was 5 or 6 bp [5][16][17][21].
many different xrs are targeted
Alignment of a number of different insertion sites of ISAba32 copies (78 different from a total of 128 insertions) showed that ISAba32 can target different xrs sites, as judged by the variation in their central regions (Fig.IS1202.11 A). An additional example was identified for ISAjo2 where 3 copies were inserted next to 3 different xrs in Acinetobacter baumannii plasmid pAF-401 (Fig. IS1202.10 Bi). This demonstrates that different ISAba32 subgroup members can target a number of different xrs sites.
Further alignments of a particular IS abutting particular xrs sites (Fig. IS1202.10 C) showed that the central regions, where xrs recombination takes place, are not conserved, and the XerC and XerD binding sites are not identical. This argues against a direct recognition of the xrs sequence by the IS transposase.
ISAba32 subgroup members target chromosomal xrs (dif sites)
A number of chromosomally located insertion sites were identified (e.g. Fig. IS1202.12). The figure shows a cumulative chromosome GC skew plot indicating positions of the replication origin and terminus [22]. Most of these insertion sites are next to chromosomal dif sites located close to the replication terminus indicating that insertion can target xrs acting in chromosome dimer resolution. The examples shown are: ISBcen27 present in two of the three Burkholderia cenocepacia MC0-3 chromosomes and also at two positions in one of the Burkholderia cenocepacia PC184 Mulks chromosomes; tISKpn21 which occurs in several Klebsiella and Serratia strains; as well as other IS in Bradyrhizobium and in a number of chromosomes of various Acinetobacter [4][1].
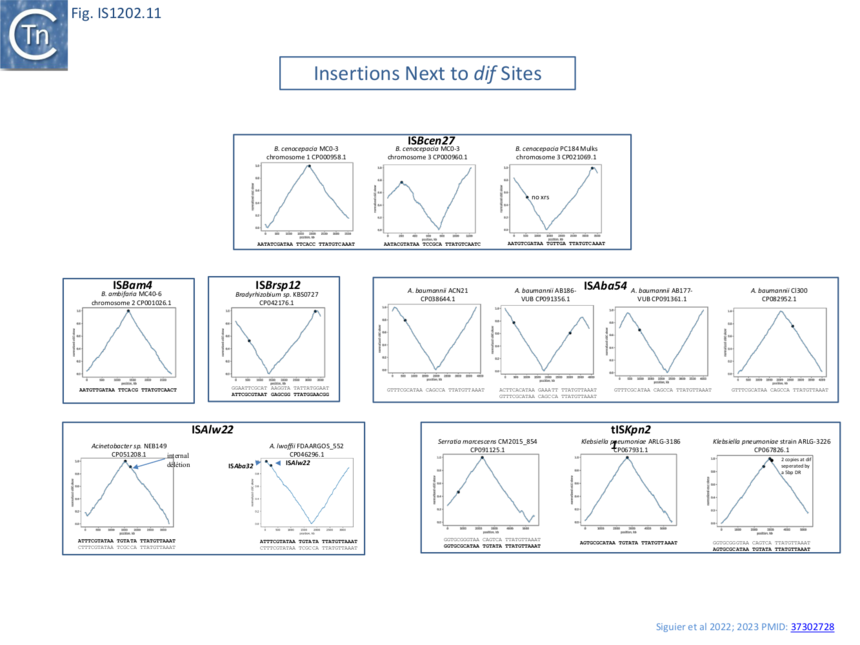
Sequential multiple targeted insertions at xrs
A number of xrs sites are abutted by several IS together with their accompanying DR (Fig. IS1202.10 C) indicating that the xrs serves as a target for successive IS insertions. This is the case for K. pneumonia ARLG-3226 which carries two tandem tISKpn21 copies at dif (although the distal tISKpn21 copy is missing a flanking DR) (Fig. IS1202.10 Civ) in addition to a third copy at an xrs site at some distance. A second example is Klebsiella pneumoniae isolate 307 plasmid P1 (OX030709) (Fig. IS1202.10 Cv) which carries 4 tISKpn21 copies each separated by the same 5 bp DR. If these insertions were targeted to the xrs site, it implies that the xrs proximal IS copy was the last to arrive since the xrs distal copy would not have targeted xrs if it arrived after the proximal copy. The third example from an unnamed plasmid in Acinetobacter junii strain ZM06 (CP077416) is more complex (Fig. IS1202.10 Cvi): there is one copy each of ISAba54 and ISAju2 separated by the 5 bp DR. There is also a second copy of ISAbA54 inserted next to a second xrs with a 5 bp DR whose sequence is different from the others (marked *). Note that there is a third non-contiguous xrs copy in this plasmid. This does not have an adjacent IS insertion. This example shows that different IS can target the same xrs and the structure again implies that the xrs-proximal IS arrived last.
Targeting mechanisms
The mechanism of ISAba32 subgroup xrs targeting is at present a matter of speculation. In view of the sequence diversity of the xrs sites targeted, it seems unlikely that ISAba32 subgroup transposases directly recognize DNA sequence. Targeting could be the result of direct transposase interactions with the XerC and or XerD proteins themselves or to a direct recognition of xrs architecture. Neither is it clear why insertion is directional i.e. that it is always IRL which abuts the xrs XerC arm: clearly IRR is less well conserved than IRL, particularly in the internal region (Fig. IS1202.8, Fig. IS1202.9 A, B, C).
One possible advantage of targeted insertion to xrs sites is that insertion could increase expression of a downstream gene either by forming a hybrid promoter [23](see TnPedia) or by providing a mobile promoter (e.g. ISEcp1; [24] IS1380 family). The IS orientation with respect to neighboring orfs is, however, often not always compatible with this. Another possibility is that they are a safe haven as in the case of insertion of the Tn7 transposon directed by an attTn7 sequence downstream from the highly conserved glmS gene (see [25]).
It is important to point out that some xrs identified in enterobacteria are flanked on their XerC-side by specific regions (called 'accessory regions') containing binding sites for various accessory proteins which serve an architectural role and control XerCD-mediated recombination [26][27][28]. As a particular DNA structure, accessory regions might be targeted by the IS. We do not know at present whether the targeted xrs possess such flanking elements that have so far been described only in enterobacteria.
In addition, targeting may inactivate possible accessory region-mediated control, damaging dimer resolution at these sites. However, the fact that ISs target plasmid-borne pdif-cassettes and chromosome dif sites, neither of which is predicted to use this kind of control, argues against this possibility.
Alternatively, the IS could act as accessory sites themselves, facilitating formation of the appropriate topology required for recombination. That the targeted xrs sites in cassettes are probably active is supported by the observation that they are recognized by Acinetobacter baumannii XerC and XerD in vitro.(P. Rouseau pers. Comm.)
Transposition Mechanism
Little is known concerning the transposition mechanism of this IS family. However, there are two reports in which circular IS copies have been identified. Hudson et al [29] identified circular copies of tISKpn21, a member of the ISAba32 subgroup, during analysis of the antibiotic resistance genes of a clinical Klebsiella pneumoniae carbapenem resistant isolate carrying the metallo β-lactamase, bla-NDM-1. MiSeq reads were found where tISKpn21 ends were linked, and separated by 5-bp identical to the direct repeat. PCR was used to rule out that this was due to tandem tISKpn21 copies in the host genome. This product is typical of the circular intermediates generated by a number of IS families by a mechanism called copy-out-paste-in [30] in which one IS end attacks the other, several base pairs into the flanking donor DNA.
Moreover, the circle appeared to be derived from a resident plasmid copy of tISKpn21 which has different 5 bp target flanks (possibly resulting from inter IS recombination): Only the left end flank was observed between the IS ends in the circle, suggesting that the right end preferentially attacks the left during circularization (rather than the left end attacking the right as stated by these authors)(see [31][32]). The second example was described by Nielsen et al [33] in a study of Sphingobium herbicidovorans MH in which they identified a circular form of an IS related to IS1202 with abutted IRL and IRR ends. Unfortunately, the DNA sequence of the collection of circles is not available in an assembled form and it is therefore not possible to identify the IS1202-related IS.
It should be noted that in IS which have adopted the copy-out-paste-in mechanism, there is generally an outward-facing -35 promoter element located in the right end and an inward-facing -10 element in the left end. This results in the temporary formation of a strong promoter in the circular intermediate which permits high levels of transposase expression (for review see [30]). Inspection of the IR sequences suggests that this may be case for tISKpn21 and may be a general property of the ISAba32 subgroup.
It remains to be seen whether the copy-out-paste-in transposition pathway is a general mechanism adopted by the entire IS1202 family.
Aknowledgements
This chapter is based on the results of Siguier et al 2022, 2023 [4][1] and the input of P. Siguier, P. Rouseau and F. Cornet is gratefully acknowledged.
Bibliography
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 Siguier P, Rousseau P, Cornet F, Chandler M . A subclass of the IS1202 family of bacterial insertion sequences targets XerCD recombination sites. - Plasmid: 2023 Jun 9, 127;102696 [PubMed:37302728] [DOI]
- ↑ Morona JK, Guidolin A, Morona R, Hansman D, Paton JC . Isolation, characterization, and nucleotide sequence of IS1202, an insertion sequence of Streptococcus pneumoniae. - J Bacteriol: 1994 Jul, 176(14);4437-43 [PubMed:8021229] [DOI]
- ↑ 3.0 3.1 Siguier P, Gourbeyre E, Chandler M . Bacterial insertion sequences: their genomic impact and diversity. - FEMS Microbiol Rev: 2014 Sep, 38(5);865-91 [PubMed:24499397] [DOI]
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 4.15 4.16 4.17 4.18 4.19 Siguier P, Rousseau P, Cornet F, Chandler M. A subclass of the IS 1202 family of bacterial insertion sequences targets XerCD recombination sites. BioRxiv. 2022 Dec 17
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 Harmer CJ, Pong CH, Hall RM . Insertion sequences related to ISAjo2 target pdif and dif sites and belong to a new IS family, the IS1202 family. - Microb Genom: 2023 Mar, 9(3); [PubMed:36880881] [DOI]
- ↑ Siguier P, Gagnevin L, Chandler M . The new IS1595 family, its relation to IS1 and the frontier between insertion sequences and transposons. - Res Microbiol: 2009 Apr, 160(3);232-41 [PubMed:19286454] [DOI]
- ↑ Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . Basic local alignment search tool. - J Mol Biol: 1990 Oct 5, 215(3);403-10 [PubMed:2231712] [DOI]
- ↑ Salter SJ, Hinds J, Gould KA, Lambertsen L, Hanage WP, Antonio M, Turner P, Hermans PWM, Bootsma HJ, O'Brien KL, Bentley SD . Variation at the capsule locus, cps, of mistyped and non-typable Streptococcus pneumoniae isolates. - Microbiology (Reading): 2012 Jun, 158(Pt 6);1560-1569 [PubMed:22403189] [DOI]
- ↑ Chamoun S, Welander J, Martis-Thiele MM, Ntzouni M, Claesson C, Vikström E, Turkina MV . Colistin Dependence in Extensively Drug-Resistant Acinetobacter baumannii Strain Is Associated with ISAjo2 and ISAba13 Insertions and Multiple Cellular Responses. - Int J Mol Sci: 2021 Jan 8, 22(2); [PubMed:33430070] [DOI]
- ↑ 10.0 10.1 Crozat E, Fournes F, Cornet F, Hallet B, Rousseau P . Resolution of Multimeric Forms of Circular Plasmids and Chromosomes. - Microbiol Spectr: 2014 Oct, 2(5); [PubMed:26104344] [DOI]
- ↑ 11.0 11.1 11.2 Siguier P, Gourbeyre E, Varani A, Ton-Hoang B, Chandler M . Everyman's Guide to Bacterial Insertion Sequences. - Microbiol Spectr: 2015 Apr, 3(2);MDNA3-0030-2014 [PubMed:26104715] [DOI]
- ↑ Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D . Highly accurate protein structure prediction with AlphaFold. - Nature: 2021 Aug, 596(7873);583-589 [PubMed:34265844] [DOI]
- ↑ Richardson JM, Dawson A, O'Hagan N, Taylor P, Finnegan DJ, Walkinshaw MD . Mechanism of Mos1 transposition: insights from structural analysis. - EMBO J: 2006 Mar 22, 25(6);1324-34 [PubMed:16511570] [DOI]
- ↑ Spínola-Amilibia M, Araújo-Bazán L, de la Gándara Á, Berger JM, Arias-Palomo E . IS21 family transposase cleaved donor complex traps two right-handed superhelical crossings. - Nat Commun: 2023 Apr 22, 14(1);2335 [PubMed:37087515] [DOI]
- ↑ 15.0 15.1
Blackwell GA, Hall RM . The tet39 Determinant and the msrE-mphE Genes in Acinetobacter Plasmids Are Each Part of Discrete Modules Flanked by Inversely Oriented pdif (XerC-XerD) Sites. - Antimicrob Agents Chemother: 2017 Aug, 61(8); [PubMed:28533235]
[DOI]
- ↑ 16.0 16.1 16.2 16.3 Hamidian M, Hall RM . The AbaR antibiotic resistance islands found in Acinetobacter baumannii global clone 1 - Structure, origin and evolution. - Drug Resist Updat: 2018 Nov, 41;26-39 [PubMed:30472242] [DOI]
- ↑ 17.0 17.1 17.2 17.3 Moran RA, Liu H, Doughty EL, Hua X, Cummins EA, Liveikis T, McNally A, Zhou Z, van Schaik W, Yu Y . GR13-type plasmids in Acinetobacter potentiate the accumulation and horizontal transfer of diverse accessory genes. - Microb Genom: 2022 Jun, 8(6); [PubMed:35731562] [DOI]
- ↑ 18.0 18.1 Balalovski P, Grainge I . Mobilization of pdif modules in Acinetobacter: A novel mechanism for antibiotic resistance gene shuffling? - Mol Microbiol: 2020 Nov, 114(5);699-709 [PubMed:32594594] [DOI]
- ↑ Shao M, Ying N, Liang Q, Ma N, Leptihn S, Yu Y, Chen H, Liu C, Hua X . Pdif-mediated antibiotic resistance genes transfer in bacteria identified by pdifFinder. - Brief Bioinform: 2023 Jan 19, 24(1); [PubMed:36470841] [DOI]
- ↑ Girlich D, Bonnin RA, Dortet L, Naas T . Genetics of Acquired Antibiotic Resistance Genes in Proteus spp. - Front Microbiol: 2020, 11;256 [PubMed:32153540] [DOI]
- ↑ 21.0 21.1 Blackwell GA, Holt KE, Bentley SD, Hsu LY, Hall RM . Variants of AbGRI3 carrying the armA gene in extensively antibiotic-resistant Acinetobacter baumannii from Singapore. - J Antimicrob Chemother: 2017 Apr 1, 72(4);1031-1039 [PubMed:28073968] [DOI]
- ↑ Grigoriev A . Analyzing genomes with cumulative skew diagrams. - Nucleic Acids Res: 1998 May 15, 26(10);2286-90 [PubMed:9580676] [DOI]
- ↑ Vandecraen J, Chandler M, Aertsen A, Van Houdt R . The impact of insertion sequences on bacterial genome plasticity and adaptability. - Crit Rev Microbiol: 2017 Nov, 43(6);709-730 [PubMed:28407717] [DOI]
- ↑ Lartigue MF, Poirel L, Aubert D, Nordmann P . In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring beta-lactamase gene blaCTX-M of Kluyvera ascorbata. - Antimicrob Agents Chemother: 2006 Apr, 50(4);1282-6 [PubMed:16569841] [DOI]
- ↑ Peters JE . Targeted transposition with Tn7 elements: safe sites, mobile plasmids, CRISPR/Cas and beyond. - Mol Microbiol: 2019 Dec, 112(6);1635-1644 [PubMed:31502713] [DOI]
- ↑ Bregu M, Sherratt DJ, Colloms SD . Accessory factors determine the order of strand exchange in Xer recombination at psi. - EMBO J: 2002 Jul 15, 21(14);3888-97 [PubMed:12110600] [DOI]
- ↑ Alén C, Sherratt DJ, Colloms SD . Direct interaction of aminopeptidase A with recombination site DNA in Xer site-specific recombination. - EMBO J: 1997 Sep 1, 16(17);5188-97 [PubMed:9311979] [DOI]
- ↑ Colloms SD, Alén C, Sherratt DJ . The ArcA/ArcB two-component regulatory system of Escherichia coli is essential for Xer site-specific recombination at psi. - Mol Microbiol: 1998 May, 28(3);521-30 [PubMed:9632255] [DOI]
- ↑ Hudson CM, Bent ZW, Meagher RJ, Williams KP . Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. - PLoS One: 2014, 9(6);e99209 [PubMed:24905728] [DOI]
- ↑ 30.0 30.1 Chandler M, Fayet O, Rousseau P, Ton Hoang B, Duval-Valentin G . Copy-out-Paste-in Transposition of IS911: A Major Transposition Pathway. - Microbiol Spectr: 2015 Aug, 3(4); [PubMed:26350305] [DOI]
- ↑ Polard P, Chandler M . An in vivo transposase-catalyzed single-stranded DNA circularization reaction. - Genes Dev: 1995 Nov 15, 9(22);2846-58 [PubMed:7590258] [DOI]
- ↑ Lewis LA, Gadura N, Greene M, Saby R, Grindley ND . The basis of asymmetry in IS2 transposition. - Mol Microbiol: 2001 Nov, 42(4);887-901 [PubMed:11737634] [DOI]
- ↑ Nielsen TK, Rasmussen M, Demanèche S, Cecillon S, Vogel TM, Hansen LH . Evolution of Sphingomonad Gene Clusters Related to Pesticide Catabolism Revealed by Genome Sequence and Mobilomics of Sphingobium herbicidovorans MH. - Genome Biol Evol: 2017 Sep 1, 9(9);2477-2490 [PubMed:28961970] [DOI]