IS Families/IS110 family-new
Contents
- 1 Historical
- 2 Organization
- 3 Mechanism
- 3.1 Transposase activity: a circular transposition intermediate
- 3.2 Transposon Circles
- 3.3 Transient Promoter Formation: the circle junction
- 3.4 Insertion specificity and target secondary structures
- 3.5 Extensive Bionformatic Analysis of Target Sequences
- 3.6 Mechanism: ISPpu9 and its Regulation by RNA
- 3.7 RNA from the NCR may be Involved with Target choice and Integration.
- 3.8 A Specific Guide RNA
- 3.8.1 Identification of Specific NCR from IS621 (IS1111) with Strong Transposase Affinity
- 3.8.2 A Consensus ncRNA Structure for IS621 Orthologues
- 3.8.3 Extending the Consensus to Other Group Members.
- 3.8.4 An Invasion Model for Bridging Donor and Target Sequences
- 3.8.5 An Efficient in vitro Recombination Reaction: ncRNA Functions to Bridge Donor and Target.
- 3.8.6 Testing the Model: an in vivo Plasmid-Based Integration System.
- 3.8.7 Reprogramming Bridge RNA
- 3.9 Flexibility in IS621 Target Specificity.
- 4 Analysis of a Second IS110 Group Member: ISEc21.
- 5 Exploring Bridge RNA Secondary Structures from Other IS110 Group Members
- 6 Use in Genome Modification
- 7 Structural Analysis: the Synaptic Complex Involved in Circle integration
- 8 Mechanism Involved in the First Transposition Step: Circle Formation?
- 9 Take Home Messages.
- 10 Bibliography
Historical
IS110 was originally identified in 1985 in Streptomyces coelicolor A3(2) as an element present in a derivative of bacteriophage phiC31 carrying a selectable viomycin resistance gene. The phage was deleted for its attachment site and therefore unable to lysogenise its host. The presence of IS110 enabled the phage to integrate using homologous recombination with resident IS110 copies in the chromosome [1].
There are over 350 examples of IS110 family members from nearly 130 bacterial and archaeal species in the ISfinder database (December 2024) [2]. However, very large number of Tpases of several have been identified in various sequenced bacterial genomes although the ends of most of these elements have not been defined and are therefore not included in ISfinder. Members such as the Mycobacterium paratuberculosis-specific IS900 and IS901 and the Coxiella burnetti IS1111 [3] have been used as a highly specific marker for precise strain identification (e.g. [4][5][6][7][8][9][10][11][12][13]).
The family includes two subgroups which, it has been suggested, may represent two distinct families [14][15]: IS110 and IS1111. Members of the IS1111 sub-group are distinguished from those of the IS110 group principally by the presence of small (7 to 17 bp) sub-terminal IRs (Fig.IS110.1) and, recognized more recently, the location of relatively long non-coding regions. Perhaps one of the earliest studied IS110 group member was IS492, from Pseudomonas atalantica originally identified by its activity in extracellular polysaccharide production (eps): inactivating the gene by insertion and reactivating by excision [16][17].

Members of the family carry a DEDD transposase and, at present is the only IS family known to encode this type of enzyme. DEDD transposases are related to the RuvC Holliday junction resolvase [19]. The Tpase is closely related to the Piv and MooV invertases from Moraxella lacunata / M. bovis [20][21] and Neisseria gonorrhoeae [22][23][24] (Fig.IS110.2). Piv catalyses inversion of a DNA segment permitting expression of a type IV pilin. Intriguingly, early studies revealed that the transposase of one IS, IS621, clustered within the piv clade (Fig.IS110.2 A) and the IS carries ends with similarities to those of the 26 bp pilin gene inversion sequences [22] (Fig.IS110.2 B). Several piv-like genes (irg1-8 for invertase-related gene) were identified in Neisseria gonorrhoeae strain FA1090 [24]. None could complement either the Moraxella lacunata Piv or IS492 transposase and inactivation of all eight genes and overexpression of one copy of each failed to show an effect on pilin variation, DNA transformation or repair.
Furthermore, analysis of DNA flanking the coding sequences support the hypothesis that the Piv homologues are indeed transposases for two new IS110 family members, ISNgo2 and ISNgo3. ISNgo2 (irg3, 4, 5, 6 and 8) is present in multiple copies in N. gonorrhoeae while ISNgo3 (irg7 and also closely related to pivNM1) is found in single copy in N. gonorrhoeae and in duplicate copies Neisseria meningitidis [24]. However, neither has yet been formally shown to transpose. Care should therefore be exercised in distinguishing between IS110 family transposases and functional piv genes.
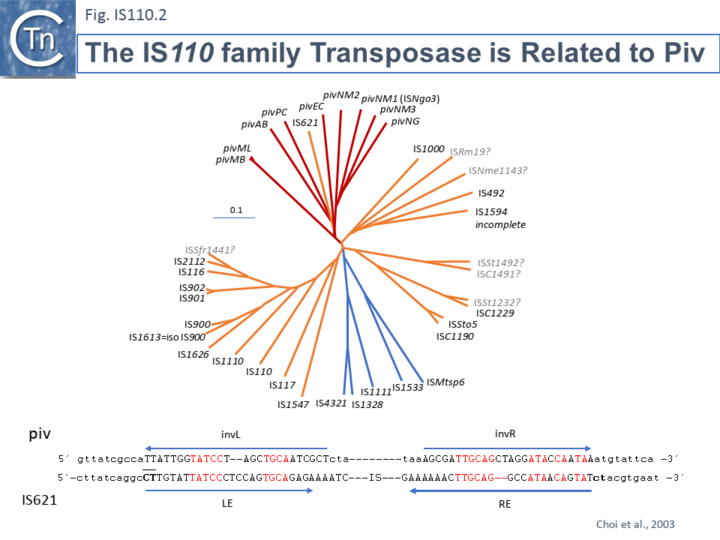
One major difference in the organization of IS110 family members and the inversion systems is that, in the piv system, the recombinase is located outside the invertible segment, while in the IS110 family, it is located within the IS element [19]. It is interesting that the piv gene is located in a cluster of IS elements in the IS110 group (Fig. IS110.2, 3A and 3B). It has been pointed out that the ends of IS621, an IS closely related to piv (Fig. IS110.2) bear some resemblance to the piv recombination site [18]; Fig IS110.2 B).
Organization
IS110 and IS1111 Subgroups Based on Transposase Sequences.
Although the Tpases of the IS110 and IS1111 groups are very similar, more detailed analysis of those in the ISfinder library showed that they generally separate into two distinct groups delineating the IS110 members (orange segment in the figure) from those of the IS1111 group (blue segment in the figure) (Fig.IS110.3) and a deeply branching segment containing a mixture of both IS subgroups (green segment in the figure), and observation confirmed by Siddiquee et al., 2024 [25]. It is possible that the few IS110 elements found within the IS1111 group and the IS1111 elements within the IS110 group have been misclassified. A similar pattern was observed in a library of transposases from over 1000 family members including members of the ISfinder collection and members extracted from public databases (Fig.IS110.3B; [26][27]). The position of piv is indicated in the figure, again, close to IS621.
Clearly, in addition to the major subgroup division, IS110 and IS1111, of this family, each contains additional deep branching clusters [25] more clearly shown in the analysis of Durrant et al., [26][27]; (Fig.IS110.3B).
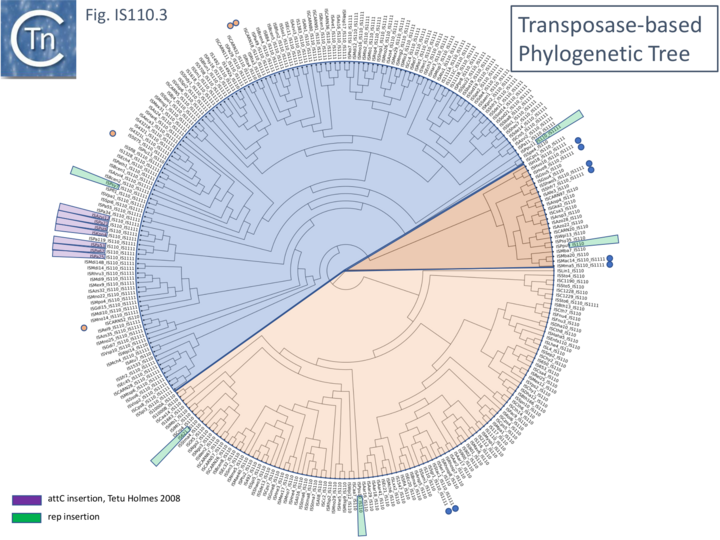
Fig. IS110.3B. A phylogenetic tree based 1,054 IS110 family recombinase sequences. The small circles indicate those family members cataloged in the ISfinder database [2]. The segments are colored as in Fig. IS110.3 A: blue, IS1111 group ; pale orange, IS110 group; darker orange segment indicates a clade with a mixture of both. Modified from Durrant et al [26].
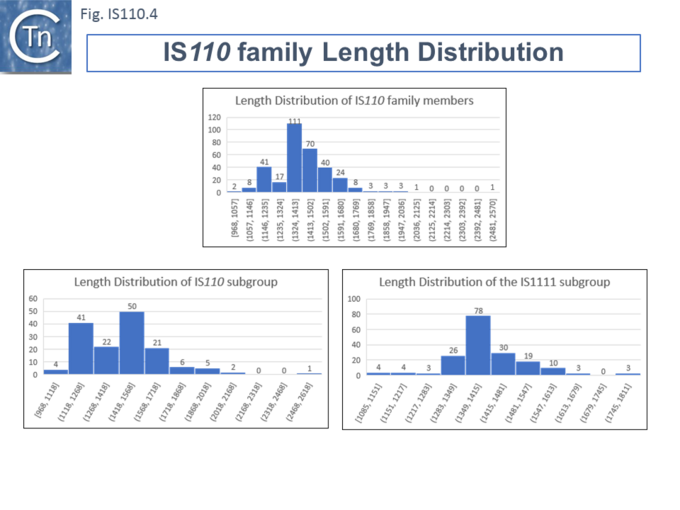
Length Distribution.
Members (Fig.IS110.4) vary between 1136 bp and 1558 bp, with most clustered in the 1450 bp size range. The length distribution of the IS110 group is more disperse than that of the IS1111 group. The organization of IS110 family members is quite different from that of IS with DDE transposases: they do not contain the typical terminal IRs of the DDE IS and do not generally generate flanking target DRs on insertion. This implies that their transposition occurs using a different mechanism to that of DDE IS.
Direct Target Repeats, DR and the Problem of Defining the Ends
Some family members have been reported to generate small Direct Repeats (DRs) while others do not (e.g. Gómez-García et al [30] and [18]). However, in most cases where flanking DR occur, the data can be interpreted to show that one DR copy is present in the target while the second copy belongs to the IS and is transmitted via a circular transposition intermediate suggesting that integration is sequence-targeted. The fact that identification of IS110 and IS1111 ends is problematic due to the absence of terminal inverted repeats might also confound the question of the presence or absence of DR. The most conclusive way to identify the IS ends would be to compare empty and occupied sites or to determine the DNA sequence across the junction formed by the abutted IS ends of the circular DNA intermediate (see below ….). This is rarely undertaken. In this light, it should be noted that many of the IS110 family in ISfinder may have incorrect ends and require readjustment.
Subterminal inverted repeats.
Partridge and Hall [31] observed that a number of IS1111 subgroup members carry long sub-terminal inverted repeats (IRst) (Fig. IS110.5 Left ) of 11 to 13 bp. These were located at approximately 6-7 bp from the left and 3-4 bp from the right end and were quite similar. As for other IS, these sequences might be expected to be recognized and bound by the transposase. IS110 group members do not carry these long IRst. However, when Durrant et al [26][27] undertook a covariance analysis of a number of IS1111 and IS110 group members, they not only observed the long IRst in the IS1111 group but also revealed very short IRst in the IS110 group (Fig. IS110.5 Right).
Fig. IS110.5 Subterminal Inverted Repeats. Left: Long Subterminal inverted repeats identified in a number of IS1111 group members [31]. Right: Results of Covariation analysis of IS110 donor sequences identified a short subterminal IR. Target and donor sequences were analysed using a covariation analysis in a large sequence library; target sequences showed no detectable covariation signal; donor sequences showed a prominent 3-base covariation signal corresponding to a LE ATA tri-nucleotide and an RE TAT tri-nucleotide. The features of both IS ends of IS110 and IS1111 group elements shown using the actual sequences of IS621 (IS110) and IS1111A (IS1111) as examples. The IS is shown as a yellow box with a purple arrow indicating the transposase orf and it direction od expression. Left (LE) and right (RE) ends are indicated. Target DNA is shown in green, the core sequences involved in recombination (see later) in blue and the subterminal inverted repeats in red [26].
Non Coding Region (NCR).
Unlike many IS families, the transposase orf does not occupy the entire IS length. Members of the IS110/IS1111 family contain a non-coding region (NCR). This was noted for ISPpu9, an example which is clustered with both IS110 and IS1111 related IS (Figs. IS110.3A and B), to include both upstream and downstream NCR regions [30].
However, there appears to be a distinction between the IS110 and IS1111 group in this respect. For the IS110 group, the NCR is generally upstream of the tnp orf while in the IS1111 group it is located downstream [25][26][27]). A number of examples are shown in Fig.IS110.6A. Although most conform to the IS110/IS1111 pattern, several such as IS621, ISRta3, ISHvo9, ISAzo22 and ISPpu9, exhibit both the upstream and downstream regions (Fig.IS110.6A) although in the case of ISPpu9, the downstream NCS is due to the presence of an ISPpu9 MITE (Fig. IS110.6B).
Fig IS110.6A
MITEs and the case of ISPpu9
In the case of one of these, ISPpu9, the downstream region results largely from an extension which appears to be a diverged defective ISPpu9 copy and includes a junction of the right (RE) and left (LE) ends separated by a characteristic AG dinucleotide (a characteristic dinucleotide which flanks ISPpu9 insertions [30]. This was identified from an analysis of the Pseudomonas putida KT2440 genome. Pseudomonas putida KT2440 carries seven copies of ISPpu9 each inserted site-specifically into one of the more than 900 35bp highly conserved REP sequences [30]. The insertions are flanked by a 2 bp dinucleotide (5’AG 3’). They found two types of ISPpu9 derivative with intact transposases (Fig. IS110.6Bi and ii): two ISPpu9 copies which we will call wildtype (wt; Fig. IS110.6Bi) and five copies of the ISPpu9 catalogued in ISfinder (Fig. IS110.6Bii). Moreover, three copies of a third (defective) ISPpu9, devoid of the tnp gene but including both left (LE) and right (RE) ends were also identified (Fig. IS110.6Biii).
These were called “orphans”. They are in fact IS110 family MITEs. The catalogued IS carries an extension on the right which includes an abutted right and left end separated by an AG dinucleotide (Fig. IS110.6Bii). This resembles the junction expected to form in a circular transposition intermediate (see: Transposon Circles) while the region downstream is similar to, but diverges from, the non-coding region upstream of the transposase gene (Fig. IS110.6Bi). These similarities and differences between the upstream NCR and the sequence of the “orphan” were pointed out by Gomez-Garcia et al [30]. It produces an RNA which the authors called Ssr9 (see Mechanism: ISPpu9 and regulation by RNA below) which was also identified in other Pseudomonas putida strains: in Pseudomonas sp KBS0802, immediately downstream of the tnp genes in five cases with one in tandem and three independent copies; in Pseudomonas putida NCTC13186, immediately downstream of six of the seven tnp copies with an additional ssr9 gene in tandem in two of these, and four independent copies, two of them in tandem, in different genomic locations. This suggested that theISPpu9def copies could transpose independently (“detach from the tnp gene”; Gomez-Garcia et al [30]).
Fig. IS110.6B. ISPpu9 Types found in the Pseudomonas putida KT2440 Genome. The transposable elements are represented by yellow horizontal boxes and transposase genes by horizontal purple arrows indicating the direction of expression. The left (LE) and right (RE) ends are represented by grey boxes. i) ISPpu9. The red panel above shows the degree of similarity with the right end of the longer ISPpu9 derivative which includes the ISPpu9 MITE. Ii) ISPpu9 including a short MITE. Iii) The MITE which has also been called an “orphan” [30].
Transposase Coding Sequence.
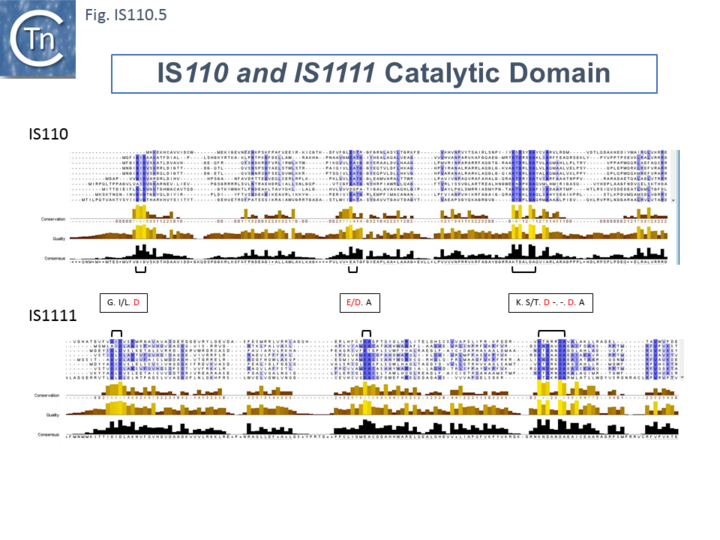
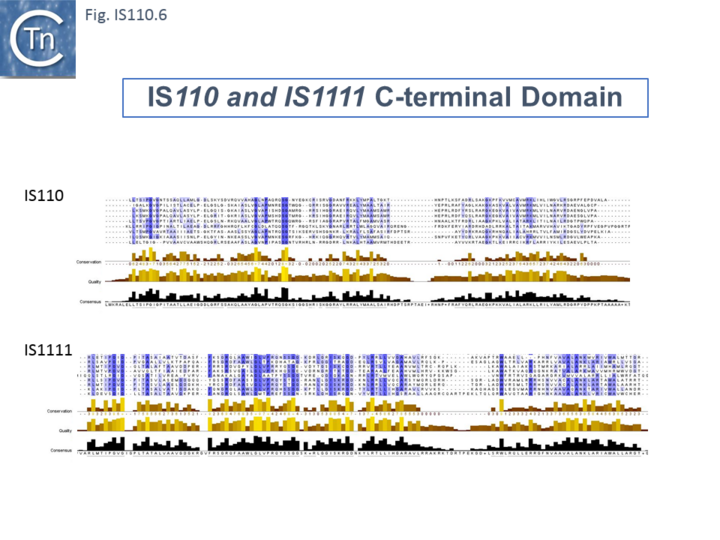
The single long, relatively well conserved, transposase reading frame shows some clusters of conservation within the N- and C-terminal portions. One characteristic which distinguishes IS110 family members from all other elements whose Tpases exhibit a predicted RNase fold is that the predicted catalytic domain of their DEDD Tpases is located N-terminal to the DNA binding domain [22][32] (Fig.IS110.1). In the DDE Tpases it is generally located downstream towards the C-terminal end of the protein. The alignment shown in Fig.IS110.5, based on 149 IS110 and 187 IS1111 group members, shows that the N-terminal catalytic domain of both IS110 and IS1111 groups share significant identities.
The probable C-terminal DNA binding domains of the two groups vary somewhat from each other (Fig.IS110.6). Those of the IS1111 group show significant conservation compared with IS110 group members, perhaps reflecting the different types of ends carried by each group.
It has been pointed out that, while the C-terminal transposase ends are somewhat variable, both the IS110 and IS1111 subgroups show a conserved SG residue [25]; [26]). Moreover, as can be seen from Fig. 110.6, the shared conserved residues are not restricted to SG but are somewhat more extensive.
Predicted Transposase Structures
Siddiquee et al., [25] used AlphaFold to predict the structure of several IS110 family transposases including ISEc21 (IS110 group) and ISEc11 (IS1111 group). Not unexpectedly, both these transposases are remarkably similar (a major reason to have grouped them into a single family in ISfinder) and also closely correspond to the structure obtained from cryo-em [33]; Fig.IS110.38 and 40). AlphaFold predicted the three domain structure composed of an N-terminal RuvC-fold catalytic domain carrying the DEDD amino acid cluster (Fig. IS110.7), a C-terminal domain carrying the catalytic Serine (Fig. IS110.7) and a coiled coli domain composed of two α-helices separated by a variable linker region. Both dimer and tetramer structures were also predicted and proved to be remarkably accurate.
Mechanism
Transposase activity: a circular transposition intermediate
It has proved difficult to determine the activity of these Tpases in detail in vitro. Transposition of IS with DEDD Tpases may be unusual and involve Holliday Junctions (HJ) intermediates [34] which must be resolved using a RuvC-like mechanism [35]. This type of recombination would be consistent with the close relationship between DEDD Tpases and the Piv/MooV invertases which presumably resolve HJ structures during inversion [36]. The difference in domain organization between the DEDD and DDE Tpases reinforces the idea that the two IS types possess a different transposition mechanism.
Few data are available concerning enzymatic activities of the putative Tpases of this family of elements: the IS900 Tpase was detected by immunological methods in the Mycobacterium paratuberculosis host [37] and the purified IS492 Tpase was reported to exhibit DNA cleavage activity specific for the ends of the element (Perkins-Balding and Glasgow, pers. comm.).
Subsequently, other IS110 transposases have now been purified including those of ISEc11, ISKpn4, ISPa11, ISPst6 (IS1111 group) and ISEc21 (IS110 group) [25] and IS621 [26] which all co-purify with, or have high affinity to, an RNA species (see XXXX below).
Transposon Circles
Several members of this family from both the IS110 and IS1111 groups produce double strand circular transposon copies. This was first demostrared for IS492:[38][39] and subsequently for a number of others (e.g., ISPa11 [15]; ISEc11 [40] ; IS117 [41][42]; IS1383 [43], IS1575 and IS4321 [44], ISKpn4 [45], and ISEc21 [46].
It should be noted that although, like other IS families, such circles are almost certainly transposition intermediates and, where examined, their formation requires transposase expression, IS110 family transposon circles could simply be generated by site-specific recombination rather than by the copy-out-paste-in mechanism adopted by families such as the IS3 family.
That the circles may be transposition intermediates was suggested by the observation that Streptomyces coelicolor IS117 was initially demonstrated in a circular form which integrates at a frequency two orders of magnitude higher than when cloned as a "linear" copy [41]. For IS117/IS116 (IS110) [41][47][48][49], IS492 (IS110) [38][50], IS1383 (IS1111) [43], ISEc11 (IS1111) [40], IS4321/IS5075 (IS1111) [15] and ISPa11 (IS1111) [15], DNA fragments carrying abutted IS ends were detected by PCR analysis in vivo and the structures confirmed by nucleotide sequencing. Their appearance was dependent on an intact Tpase gene and their nucleotide sequence is consistent with the formation of a circular form of the element.
Henderson et al, 1989[41] were perhaps the first to suggest that this family used site-specific recombination to transpose. IS117, originally identified as a “mini” circle shows a 2/3 base pair identity between the circle junction and its specific site of insertion into the host chromosome [41][47][48] (Fig.IS110.7). Transposition was often found to result in tandem dimer inserts, behavior which might indicate some type of rolling circle insertion mechanism such as observed in the case of the IS91 family elements.
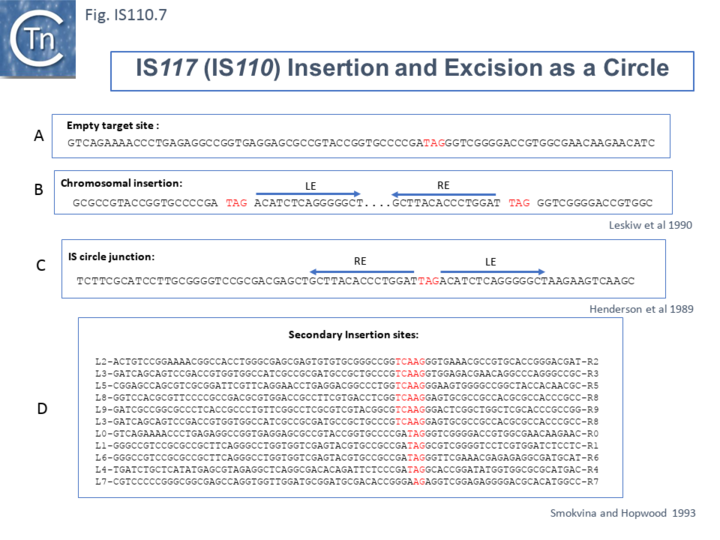
Another member of the IS110 group, IS492, clearly undergoes Tpase dependent precise excision to regenerate a functional eps gene in Pseudomonas atlantica (Fig.IS110.8 A). The inserted IS copy is flanked by 5 bp directly repeated sequences (5’-CTTGT-3’) (Fig.IS110.8 B). The circle junction carries a single copy of this sequence (Fig.IS110.8 C) as does the empty target site. This suggested that one copy is carried by the IS and is required for activity. Sequential deletion of the ends of (Fig.IS110.8 D) clearly showed that the pentanucleotide and/or sequences immediately upstream were required for excision. On the other hand, a sequence 5’-GTTT-3’ located upstream in those insertions analyzed (Fig.IS110.9) was not required for excision. It is possible that they are needed for circle integration.
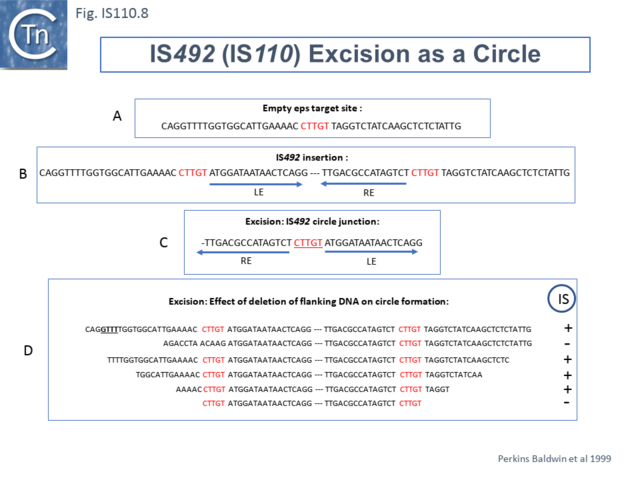
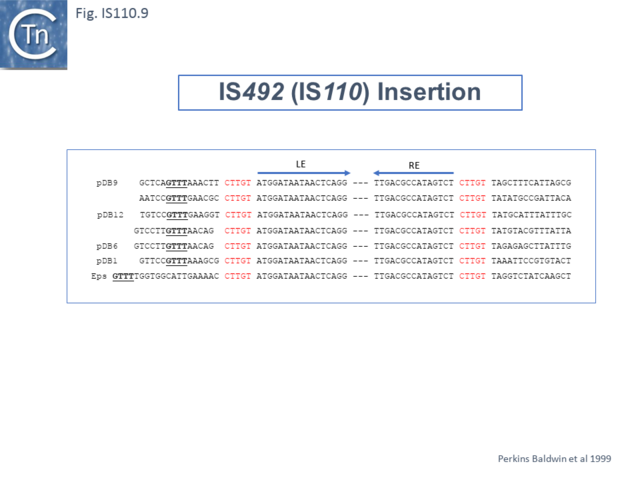
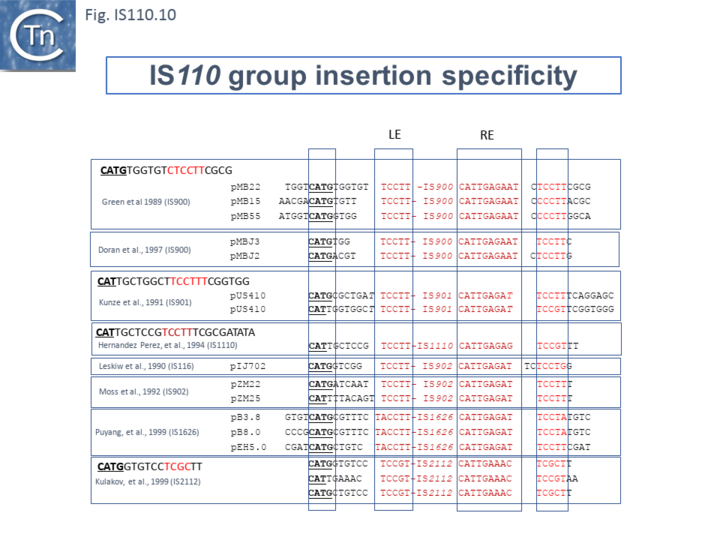
Similar flanking sequences have also been identified in insertions of IS900, IS901, IS902, IS116, IS1110, and IS2112 (Fig.IS110.10) and IS621 was also shown to have a flanking sequence, in this case a dinucleotide, CT [22].
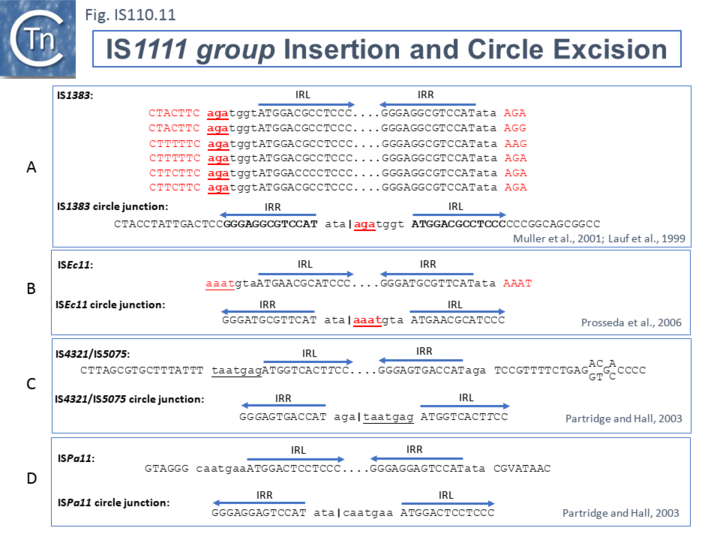
The ends of IS1111 group members differ from those of the IS110 group by including short subterminal IRs. IS1383 was identified as flanking insertions into each end of the IS5 family member, IS1384 [43][51] and was also shown to generate IS circle junctions (Fig.IS110.11 A). Like most members of this group, IRL is located further from the IS tip than is IRR. In this case IRL is preceded by the sequence 5’-agatgg-3’ (lower case indicates the IS end sequences upstream and downstream of IRL and IRR respectively). The insertions into the ends of IS1384 had occurred into a resident AG(A) sequence and excision to form the circle junction appeared to have occurred by recombination between the resident AG(A) and the terminal aga at the left end of IS1383 [43]. This this is compatible with a site-specific recombination mechanism in IS1383 transposition. A similar arrangement was observed for a second IS1111 group member, ISEc11 [40], where a flanking tetranucleotide AAAT also appeared as part of the circle junction (Fig.IS110.11 B) and it has also been argued that this is compatible with a site (sequence)-specific recombination transposition mechanism [40]. However, in two additional cases from the Hall lab, IS4321/IS5075 and ISPa11, no such “micro-homologies” were detected [15] (Fig.IS110.11 C and D). However, it should be noted that transposon circles are generated in vivo and analyzed by PCR. Since there may be a number of copies of the IS in the host genome, this might compromise the sequence of the PCR product.
The number of fully studied examples of IS1111 group members is limited, it is possible that the flanking “micro-homologies” observed for IS1383 and ISEc11 are chance occurrences and that excision and insertion of IS1111 members is truly mechanistically different from those of IS110 group members and that their division into separate families is justified. However, for present classification, both groups are included in the IS110 family in ISfinder for convenience.
Transient Promoter Formation: the circle junction
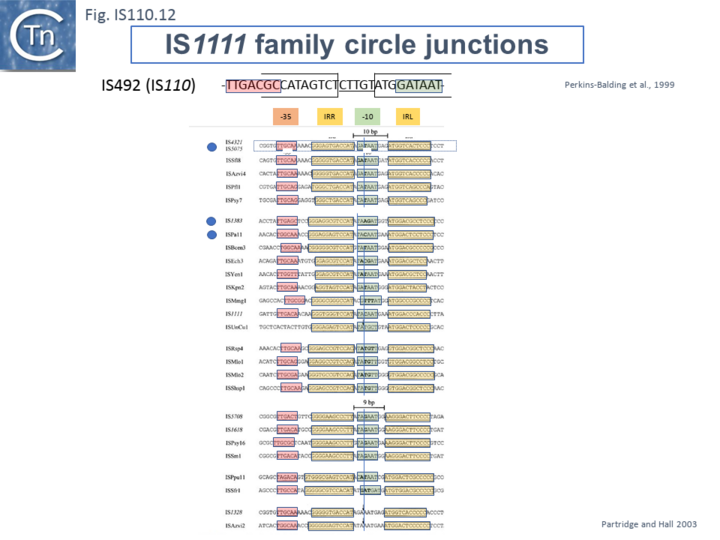
Like many other IS which use double strand circular intermediates, circle formation results in the assembly of a junction promoter formed from a -35 promoter element in the right end oriented outwards and a -10 promoter element in the left end oriented inwards [52][53][54]. For the IS110 family, this was originally identified in circular forms of IS492 [38] (Fig.IS110.12). A list compiled of many IS1111 group IS [15] and in silico construction of IS circle junctions indicated that all had the capacity to generate probable promoters. Due to small variations in the distance of the subterminal IRs from the probable end of the IS, some were separated by 10 bp and some by 9 bp. A notable observation is that while the -35 promoter elements are located entirely within the right IS end, the -10 promoter element was not located entirely within the left end but was composed of sequences from both the left and right ends and was only assembled on circle formation. However, unlike the IS492 junction promoter which appears to be significantly stronger than the lacUV5 promoter [38] and the junction promoters of ISEc11 and a naturally occurring derivative, ISEc11p which are also functional [40], few of these have been examined for activity.
Insertion specificity and target secondary structures
The particular insertion specificities of the IS110 family has been mentioned in the context of the mechanism of transposition and is often one factor in making definition of the IS ends difficult. However, one characteristic of insertion of this family of IS is that they offen prefer sequences with the propensity to form secondary structures. This is consistent with the fact that the transposases are similar to the RuvC and the RuvC endonuclease is involved in resolving branched Holliday junctions during recombination (e.g.[55]).
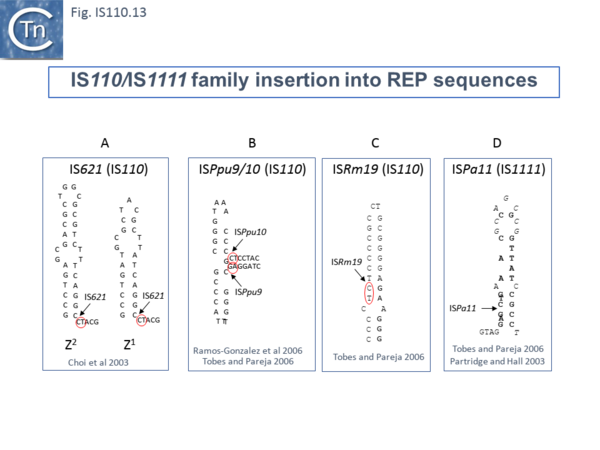
For example, IS621 insertions were observed to be flanked by a CT dinucleotide [22]. On further examination this was shown to be a dinucleotide located at the foot of Rep sequences in the host Escherichia coli genome (Fig.IS110.13 A). REP sequences are small Repeated Extragenic Palindromic sequences often present in many hundreds of copies in bacterial genomes and which play a variety of structural and regulatory roles [56][57][58][59][60][61][62]. Both Z1 and Z2 Rep [57][58][59] sequences are used as targets and all 10 copies of IS621 in the E. coli ECO28 genome were found in this position in resident Rep sequences [22].
There are at least six other examples of this type of “structural” insertion specificity (Fig.IS110.2). All 8 copies of ISPpu10 were identified in short REP sequences of Pseudomonas putida KT2440 [63][64] and a cloned ISPpu10 derivative was shown experimentally to transpose into this REP target [63] (Fig.IS110.13 B). Eight (of 8) copies of a related IS, ISPup9, were identified in the same REP sequence at the same position but inserted in the opposite orientation (i.e. on the opposite strand)[65] (Fig.IS110.13 B) while 4/4 examples of ISRm19 were identified in a REP sequence of Rhizobium meliloti (Fig.IS110.13 C). Similarly, ISPa11 of the IS1111 group inserts specifically into a Pseudomonas aeruginosa REP (6 examples) [65] and one example from Partridge and Hall [15] (Fig.IS110.13 D).
Two types of Insertion have been described [65]. In type 1, the IS inserts at the same position within the REP whereas type 2 insertions occur adjacent to a REP. Most IS110 family members exhibit type I insertion patterns in all examples identified. However, one IS, ISPsy7 exhibited type II insertion pattern but only in 6/10 examples and a second unspecified IS from Neisseria meningitidis MC58 was also reported to exhibit a type II pattern in 3/5 cases examined [65]. It is possible that this N. meningitidis IS is the same as that described by Skaar et al. [24].
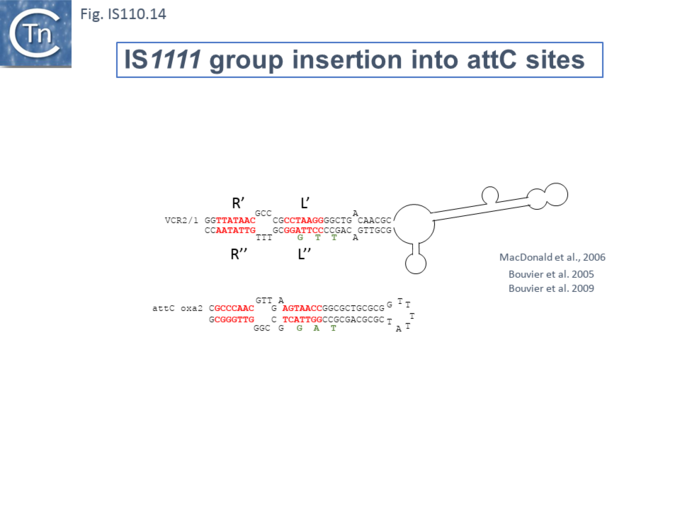
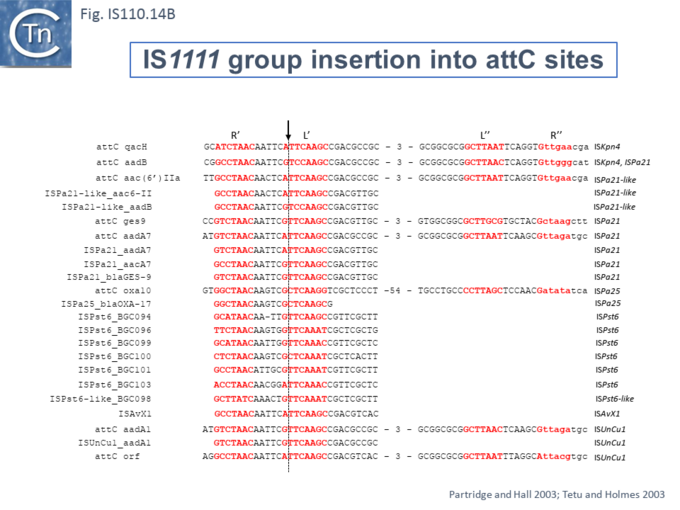
At least six different members of the IS1111 subgroup (ISKpn4, ISPa21, ISPst6, ISUnCu1 = ISPa62, ISAvX1 = ISAzvi12 and ISPa25) show a preference for another type of target which can assume a structured configuration, the attC sequences of integrons [66][67]. IS which insert into attC sequences are grouped into a specific clade (Fig.IS110.2) [66]. The integron attC is central to integration of circular integron cassettes [68] and had been called “59 base pair element” [69] but can vary considerably in length [70]. Studies from the Mazel lab have shown that attC sequences can form foldback structures (Fig.IS110.14 A) with imperfect matches in which extrahelical bases are involved in driving the direction of the excision and integration reactions [68][70][71][72]. Integration of IS1111 group members appears to occur at a specific position on these attC foldback sequences (Fig.IS110.14 B).
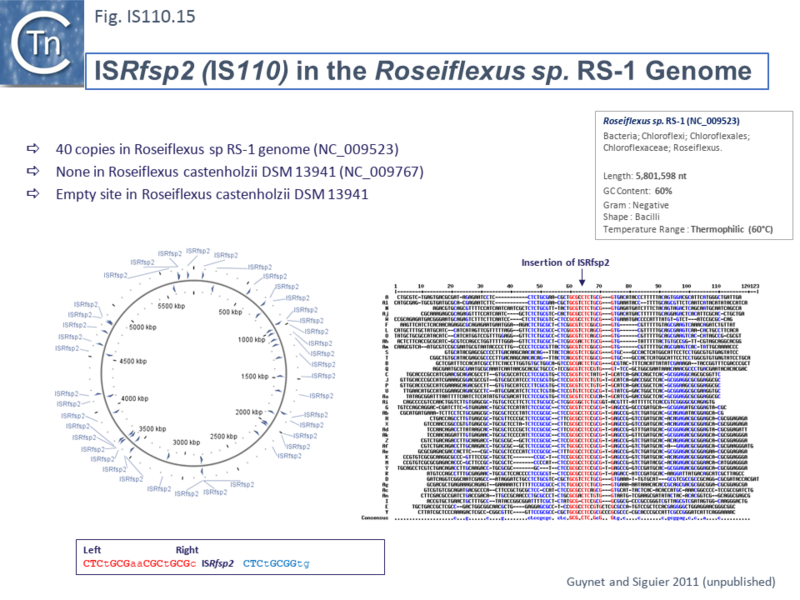
Other IS of this family also appear to insert into conserved target sequences: IS1533 occurs in 84 copies in Leptospira borgpetersenii and inserts into a partially conserved sequence (ttAGACAAAA [IS1533] TATCAGagcc-gtct--aaa); ISRfsp2 from Roseiflexus sp RS-1, present in 40 copies in the host genome, is flanked by the sequence, CTCtGCGaaCGCtGCGc [ISRfsp2] CTCtGCGGtg (Fig.IS110.15) while ISMpa1 from Mycobacterium avium subsp. Paratuberculosis is flanked by the consensus CCAGN0–1CTA [ISMpa1] GCCN0–6GCCG [73].
Extensive Bionformatic Analysis of Target Sequences
Siddiquee et al.,[25] undertook an extensive analysis of the IS110 family members in ISfinder using a library of IS together with their flanking DNA extracted from public databases and ranked in order of abundance and number of independent insertions (https://github.com/ AtaideLab/Targets/31). The different IS were found to occur with a very large range of frequencies. A number were represented only once in the library while others from both IS110 and IS1111 groups were present in very high numbers: some in several thousand with hundreds of unique insertion events. Among the most abundant were IS1663 (9059 copies; 7061 insertions), ISSfl4 (2364;1735), ISNgo2 (1268; 1017), IS621 (13214; 920), and ISSep2 (3173; 898) in the IS110 group and IS1533 (4620; 1162), ISKpn43 (1190; 1145), ISPa11 (1213; 1063), IS4321 (1092; 899), and ISYen1 (1049; 830) in the IS1111 group. Other members were present in large numbers with a unique insertion site for example: ISMba20 (225; 1); ISMba7 (184; 1); ISMch6 (17; 1), ISRhosp8 (304; 1) and ISSde13 (163; 1) in the IS110 and ISSod21 (7; 1), ISSphsp16 (39; 1), ISSphsp18 (41; 1), ISStac1 (1462; 1) and ISXpo1 (23; 1) in the IS1111 group.
Analysis of these data using WebLogo revealed that the consensus target sequences with large differences between different IS in the strength and length of the conserved sequence.
WEBLOGO Examples????
Accurate Identification of IS110 Family left and right ends.
It is important to note that there are some ambiguities in a number of the ends of IS110 family members documented in ISfinder due to the absence of terminal IRs as pointed out by Siddiquee et al., [25] the most definitive method of resolving these problems would obviously be to obtain the DNA sequence of the RE-LE IS circle junction.
Mechanism: ISPpu9 and its Regulation by RNA
One of the first suggestions that control of transposition of IS110 family members might involve RNA came from studies on ISPpu9 [30] (Fig. IS110.3A and B and IS110.6B).
Asr9 and ssr9 RNA
An analysis of transcription in Pseudomonas putida [74] led to the identification of two untranslated regions (NCR) in ISPpu9 from which two small RNAs (sRNAs) are produced: one, ssr9, is located downstream of the tnp gene (tnpISPpu9) expressed from the probable defective ISPpu9 MITE-like structure (Fig. IS110.6B) in the same direction and the second, ars9 (antisense sRNA of ISPpu9), is located upstream, convergent with the transposase promoter and expressed from the opposite DNA strand (Fig. IS110.16). Asr9 was determined to be nearly 5 times more abundant than ssr9. Tnp ISPpu9 transcripts were only detected at very low levels.
Fig. IS110.16. RNA seq on genomic ISPpu9. Top: Map of ISPpu9 (yellow horizontal box) showing the transposase gene (purple horizontal arrow) and the results of RNAseq (red). The IS ends, including those of the associated MITE on the right, are indicated by grey boxes and the promoters as black arrows. Bottom: DNA Sequence of the left and right IS regions (left and right boxes respectively). Note that the right sequence contains the entire MITE. The 5’ and 3’ REP target sequences are shown in blue boxes in lower case. Left and Right ends are indicated by grey boxes LE and RE. Inverted repeats are shown as blue arrows. The left hand box shows the probable transposase -10 promoter region, the +1 transcription start together with the transposase initiation codon are shown in red as are the probable -10 and -35 ars promoter regions and the +1 transcription start. The right hand box shows the transposase termination codon, the probable -10 region of the defunct transposase and of the ssr transcript [30].
Inspection of the sequences of both ars9 (upstream) and ssr9 (downstream) indicated a significant divergence (Fig. IS110.16 and Fig. IS110.17) which presumably eliminates the ars9 promoter in the downstream ssr9 sequence although both maintained an upstream inverted repeat.
Fig. IS110.17. Sequence Differences between the ISPpu9 Left End and the Right Hand Mite. The two sequences are aligned Red characters indicate differences. Bold characters indicate various functional nucleotides including: the probable transposase (Ptnp) -10 promoter region; the +1 transcription start (missing in the MITE); -10 ssr (missing in the ISPpu9 sequence); the ssr transcription start site (missing in ISPpu9 sequence); the ars +1 (missing in the MITE); the 1-10 and -35 ars promoter (Pars9) signals (missing in the MITE); and the transposase translation initiation codon (missing in the MITE). The LE-associated inverted repeat is present in both and the more internal inverted repeat (missing in the MITE) are shown by blue horizontal arrows. The IS ends are shown as grey boxes [30].
Clearly, asr9 could act as an anti-RNA to control transcription/translation of the tnp gene. To investigate this, a series of plasmid-based Tnp-lacZ translational fusions were constructed (Fig. IS110.18). These included derivatives containing either the first two tnp codons (called 2 and 2+S, Fig. IS110.18. 1 and 2) eliminating the ars9 -35 promoter component or the first 8 (called 8, 8+S and ; Fig. IS110.18. 3, 4 and 5) which include the entire ars9 promoter (Fig. IS110.18. 3 and 5) or a copy with a mutated -35 promoter component (Fig. IS110.18. 4). The 2 and 8 tnp codon derivatives were also constructed with (Fig. IS110.18. 2 and 5) or without the corresponding downstream ssr9 promoter (Fig. IS110.18. 1 and 3).
Propagation of these plasmids in Pseudomonas putida F1 (which is devoid of ISPpu9 or associated genes) revealed that plasmids 8 and 8+S (Fig. IS110.18. 3 and 5) produced significant levels of ars9 RNA while plasmids 2 and 2+S1 and 2 (Fig. IS110.18. 1 and 2) did not. The plasmid which had a mutated -35 promoter box (Fig. IS110.18. 4), however continued to produce a low level of the RNA. Measurement of β-galactosidase activity from these plasmids in Pseudomonas putida F1 (which is naturally devoid of ISPpu9 sequences) revealed that plasmid S (Fig. IS110.18. 1) was only 25% that of construct 8 (Fig. IS110.18. 3) although the levels of lac mRNA were only 70 % lower suggesting that the major effect of ars9 RNA was on translation.
The authors propose that the tnp ribosomal binding site in the mRNA is masked by the inherent secondary structure and that interaction with ars9 RNA liberates this, facilitating TnpISPpu9 translation (Fig. IS110.18 bottom). Moreover, introduction of an ars9 gene into the chromosome of Pseudomonas putida F1 further significantly increased β-galactosidase expression from plasmid 8 (Fig. IS110.18.3). However, this expression enhancement did not occur with plasmid 2 (Fig. IS110.18.1) and the authors suggest that this could be because asr9 cannot properly hybridize with the NCR RNA of plasmid 2 possibly because the sequence between codons 2 and 8, plasmid might be important for asr9 activity by, for example, providing an initiation point for pairing. This was not further tested.
Additionally, the presence of ssr9 appeared to alleviate the effect of ars9 suggesting that this RNA, with partial identity to the upstream NCR (Fig. IS110.17), might be able to sequester ars9 thus reducing its activity. Such an interaction was detectable in vitro. This effect was observed in Pseudomonas putida F1 as a 27% lower β-galactosidase level from the 8+S plasmid than from the 8 plasmid and a 35% lower level in the Pseudomonas putida KT2440 host.
The notion that the NCR secondary structure is responsible for sequestering the translation initiation signals is supported by the observation that a number of mutations designed to disrupt or weaken the NCR secondary structure and therefore demask the ribosome binding site resulted in a large increase in β-galactosidase expression in the absence of ars9.
Using lacZ transcriptional fusions, the activities of Pars9 and Pssr9 were found to be about 3 fold higher than Ptnp and ars9 RNA was significantly more stable (half life >60 min) than ssr9 (half life ~3 min). The authors present experiments which lead to the conclusion that asr9 stability is due to its sequence and secondary structure rather than to interaction with ssr9 or the 5’NCR RNA.
It should be noted that these studies addressed “linear” IS copies and did not involve the presumed circular intermediate which is likely to generate a strong promoter due to the assembly of abutted left and right IS ends (see: Transient Promoter Formation: the circle junction). Regulation of Tnp expression among other characteristics is likely to be modified in these transposition intermediate structures.
Fig. IS110.18. Tnp-lacZ Translational Fusions. The effect of the 5’ NCR, Asr and Ssr on Transposase expression measured by translational fusions to the lacZ reporter gene. The constructions are shown as cartoons on the left. The horizontal blue arrow represents lacZ gene and the purple box shows a fusion with either the first 2 or 8 codons of the transposase. The transposase promoter, Ptnp, is included in all constructions while the ssr promoter, Pssr, is only included in constructions (2) and (5). The complete ars promoter, Pars, with its -10 and -35 is present in constructions (3) and (5) while construction (4) carries a mutated -35. These features are shown on the aligned DNA sequences to the right together with the +1 translational start for the Ars RNA (red). -10 and -35 positions are underlined and in bold. Note that the Tnp ribosome binding site (RBStnp) is boxed and ovelaps the ars – 10 promoter component. The positions in black font correspond to tnp, those in blue (boxed) to‘ lacZ , and those in gray to extra codons introduced during cloning. Positions mutated at the -35 region of promoter Pasr9 are indicated in green. The table on the right shows the relative levels of β-galactosidase produce (-/+) and the presence (+) or absence (-) or ars- or ssr-RNA. The schemas at the bottom show how pairing of asr RNA to the 5’NCR of the tnp mRNA could unfold the hybridization loop providing access of the RBStnp to ribosomes thus facilitating tnp translation.
RNA from the NCR may be Involved with Target choice and Integration.
The involvement of an RNA from the downstream NCR in determining IS1111 group insertion specificity had been suggested [29] based on comparison of ISKpn4 and ISPa25. ISKpn4 belongs to an IS1111 subgroup targeting att sites of integron cassettes (Fig. IS110.3A) and while ISPa25 also targets att sites, it belongs to an IS1111 subgroup including IS4321 and ISPa11 (Fig. IS110.3A) whose transposases have low amino acid similarity with the ISKpn4 subgroup and targets the IR of Tn21 transposons. It was noted that ISKpn4 and ISPa25 share a block of sequence similarity in the downstream non-coding region (Fig. IS110.19) and it was suggested that, as RNA, this might be responsible for target choice. More careful analysis presented here has revealed that the two IS also share blocks of similarity at the 3’ end of their transposase genes and that this results in strong amino acid conservation in the transposase itself (Fig. IS110.19). The first block of similarity carries the G..P/SG conserved residues (Fig.IS110.8).
Fig. IS110.19. Sequence Patchwork of IS1111 Group Members: ISKpn9 and ISPa25. Top: Comparison of ISKpn4 and ISPa25. The IS are shown as horizontal yellow boxes and the transposase orfs as purple horizontal arrows showing the direction of expression. Regions of strong similarity are shown as blue boxes with the IS coordinates above (ISKpn4) or below (ISPa25). The coordinates of the transposase codons for ISKpn4 are indicated between the two IS. Middle: DNA sequences of the three blocks of similarity. ISKpn4 (top lines) and ISPa25 (bottom lines in each box). Identical nucleotides are shown in black text. Bottom: Protein Sequence of the C-Terminal transposase end. The block od similarity are shown in blue (bold) and the identities are underlined.
A Specific Guide RNA
Durrant et al [26][27] extracted and aligned a large number of examples of this family from public databases (2023) (Fig. 110.3B) which greatly increased the number of family members in the ISfinder database. They observed that, compared to other IS families, members of the IS110 family exhibit some of the longest non-coding ends (NCR or Untranslated Regions, NCR) among IS families. That this is a conserved family feature is suggested by a relatively narrow length distribution (between 230 and 290 bp).
Identification of Specific NCR from IS621 (IS1111) with Strong Transposase Affinity
To further explore the mechanism involved in IS110 transposition, Durrant et al [26][27] used IS621 of the IS110 group as a model system. IS621 (Fig. IS110.2B) was first described by Choi et al [18] and comparison of a number of resident IS621 homologues in E.coli demonstrated that they insert at the foot of a REP sequence and are flanked by a CT dinucleotide (Fig. IS110.13). IS621 has both upstream and downstream NCR sequences (Fig. IS110.6A and 19). The predicted RE-LE junction of the probable IS621 circular transposition intermediate was cloned together with the tnp upstream NCR and analyzed for RNA expression in E.coli [26][27]. A prominent RNA region of approximately 170 nts was identified which appeared to originate just downstream from the junction promoter and continue until immediately before the TnpIS621 +1 codon (Fig.IS110.20).
Fig. IS110.20. IS621, the IS Circle Junction and its Transcript. Top: Map of IS621 (yellow box) showing the transposase (purple arrow) and the left and right ends (grey arrows). Bottom: the DNA sequence (black characters) across the RE-LE junction in the IS621 circular transposition intermediate. Right (RE) and Left (LE) ends are indicated within a grey box. They are separated by the CT dinucleotide (blue) which flanks the original inserted copy [18]. The junction promoter, Pjunc, -10 and -35 components are shown within yellow boxes and the transcription start site (TSS) is shown within a red box. The RNA transcript is shown as a red dotted line and the left target guide (LTG), right target guide (RTG), left donor guide (LDG) and right donor guide (RDG) sequences are shown as red characters and underlined. The transposase start codon, ATG, is shown in red.
Using purified TnpIS621 and in vitro transcribed ncRNA, it was found, using Microscale thermophoresis (MST) to determine the equilibrium dissociation constant, that the protein showed high affinity for the RNA. This is a characteristic of guide RNAs in other systems where they co-purify with their guide endonucleases (see: TnpB and its Relatives).
A Consensus ncRNA Structure for IS621 Orthologues
A consensus ncRNA (non-coding RNA) structure was then determined for over 100 IS110 orthologues using structural alignments and structural prediction software together with sequence conservation. Development of a covariance model revealed the presence of a 5’ stem-loop followed by two larger stem-loop structures each with a large internal loop (Fig. IS110.21). The first had low sequence conservation while the second was significantly more conserved.
Fig. IS110.21. Generalised Secondary RNA Structure. The consensus ncRNA secondary structure was constructed from 103 IS110 LE sequences. The predicted structure comprises a 5′ stem - loop and two large internal loops. A key is included to the right of the figure.
The strong binding of the ncRNA to the Tnp protein raised the possibility that it may favor target recognition.
Extending the Consensus to Other Group Members.
To explore this, the authors first defined the ends of a large number of IS110 elements enabling identification of their insertion sites and reconstruction of both the target sequence and the junction of the circular form. They then performed an iterative search with the structural covariance model (CM) developed for IS621 ncRNA (Fig. IS110.21) to predict ncRNA structures in the LEs of this IS collection, generated paired alignments of the ncRNAs with their corresponding target and donor (abutted LE and RE ends) using a 50bp window centered on the donor “CT” dinucleotide core, and undertook covariation analysis (2,201 donor - ncRNA pairs and 5,511 target - ncRNA pairs) detected by homology with IS621 (Seemayer et al., 2014 PMID: 25064567). This incorporated base-pairing analysis to identify stretches of these ncRNA complementary to either the top or bottom strand of the target or donor DNA. It identified possible pairings with the two internal ncRNA loops. By projecting the overall covariation pattern for the entire collection onto the model IS621 ncRNA sequence, the authors inferred that the first loop could base-pair with the target and the second to the donor junction: the 5’ side of the loop would pair with the bottom target donor strand (8-9 nts) and the 3’ end with the top strand (4-6 nts) (Fig. IS110.22A) [26][27]).
Fig. IS110.22. Co-Variance Analysis and Complementarity of ncRNA with Target and Donor. A) The analysis was carried out with 5, 511 ncRNA–target pairs (top left) and 2,201 ncRNA–donor pairs (top right). The target (left, green) and donor (right, organge) are represented vertically. The IS621 ncRNA sequence is shown below along with dot-bracket notation secondary structure predictions together with LTG and RTG sequences in green and LDG and RDG sequences in orange. Covariation scores are colored according to strand complementarity (insert bottom left): blue, high covariation and bias toward top-strand base-pairing; red, high covariation and bias toward bottom-strand base-pairing. Regions of notable covariation signal indicating base-pairing for IS621 are boxed. An extended signal for the top strand (purple lozenges) is observed and, on the IS621 sequence is indicated by the ribonucleotides UGC marked in red. The double strand target (left) and donor (right) sequences are included below showing the sequence of complementarity (boxed) Complementary nucleotides within covarying regions are highlighted in bold. The CT dinucleotide which occurs as a direct flanking repeat in the inserted IS [18] and at the circle junction is shown in blue. B) Nucleotide conservation across the predicted ncRNA. 2,715 ncRNA orthologue sequences were identified using an iterative search with the original IS621 model. Top: Nucleotide conservation represented in WebLogo format. The various secondary structure elements are indicated mapped onto the IS621 ncRNA and delimited by vertical blue lines. Stems are indicated by horizontal colored arrows. The first loop shows low sequence conservation, while the second is much more conserved. Sequence features of the bridge RNA are highlighted for clarity. From Durrant et al [26].
An Invasion Model for Bridging Donor and Target Sequences
These strong signals of co-variation and base pairing led to the idea that ncRNA bridges the target sequence and the IS circle junction during transposition and led to the “invasion” model shown in Fig. IS110.23 [26][27]. In this model both upstream and downstream loops engage and align the target and donor DNA sequences facilitating recombination at the core by the DEDD Tnp (Fig. IS110.7) presumably with the aid of the conserved serine residue located in the C-terminal domain as the nucleophile (Fig. IS110.8). The authors underline the observation that the “core” dinucleotide is included in all 4 of the base pairings (Fig. IS110.22). Thus there is an overlap between top- and bottom-strand pairings precisely at the core dinucleotide. This presumably plays a key role in the recombination (cleavage and strand exchange) reactions which was confirmed by structural studies (below).
The covariance data also suggested that the IS621 right target guide sequence (RTG) is short and that other members of the IS110 group include longer RTG (Fig. IS110.22A - note the purple extension on the Upstream Loop, Top strand). This is indicated on the IS621 sequence by the red ribonucleotides (see also Insertion in vivo)
An Efficient in vitro Recombination Reaction: ncRNA Functions to Bridge Donor and Target.
An in vitro IS621 recombination reaction was assembled to test this idea. This was composed of an in vitro-transcribed ncRNA, the purified IS621 transposase/recombinase and short, double stranded oligonucleotides containing the target and donor sequences. The reaction mixture also included NaCl and MgCl2.
Microscale thermophoresis (MST) experiments demonstrated that the ncRNA-transposase/ recombinase complex bound both donor and target DNA molecules in a sequence-specific manner. This combination of components led to the expected reciprocal DNA exchange reaction at the CT “core” site with the expected junctions as detected by appropriate PCR assays. Since the ncRNA was capable of binding both the donor IS circle junction containing abutted RE and LE as well as the target, Durrant et al [26][27] have called it a Bridge RNA (Fig. IS110.22 and 23).
Fig. IS110.23. Bridge RNA Interaction with Donor and Target. The left of the figure shows the configuration of the bridge RNA with the Target Binding Loop (TBL) which includes the left and right target guide sequences (green characters) and the Donor Binding Loop (DBL) with the left and right donor binding sequences in orange characters. Those residues which are not complementary to the donor or target sequences are shown in grey. Below (orange) and above (green) are the donor (circle junction) and target double strand DNA respectively. The “core” CT dinucleotides are marked in blue. Interaction of the TBL with the target sequence and of the DBL with the donor circle junction (right hand secton) involves unwinding of these double strand DNA segments and annealing of the LTG with the left target (LT) sequence and the RTG with the right target (RT) and of the LDG with the left donor (LD) sequence and the RDG with the right donor (RD) sequence. This facilitates recombination between the two core CT dinucleotides resulting in IS integration. Redrawn from Durrant et al [26].
Testing the Model: an in vivo Plasmid-Based Integration System.
Further support for this “invasion” model was obtained from experiments designed to reprogram either donor or target sequences. The experiments used a 2 plasmid system in vivo: one plasmid, pTarget, carried tnpIS621, the 50 bp target site (a Rep sequence) and a flanking promoter; the other, pDonor carries the RE-LE donor circle junction, the bridge RNA and a promoter-less gfp gene. Donor-target recombination places gfp under control of the pTarget promoter (Fig. IS110.24) and can be assayed by measuring fluorescence. This assay was used to monitor the effect of mutations in TnpIS621: alanine substitution of the conserved catalytic residues, DEDD, of the RuvC-like domain (Fig. IS110.7) or the recombinase domain, S, (Fig. IS110.8) abolished activity. Gfp expression was measured using a flow cytometer by scraping and resuspending colonies from a plate after co-transformation of a recipient strain with the two plasmids under standard plating conditions. In a number of cases, the plasmid sequences were also obtained to confirm the recombinant structures.
Fig. IS110.24. Gfp Activation Integration Assay. Top panel: Donor and target plasmids. Selective CmR (pTarget) and KmR (pDonor) genes are shown in red, transposase in purple with an IPTG-inducible promoter promoter (Ptnp, blue arrow), target (a Rep sequence) in dark green interrupted by the recombination point (CT dinucleotide) in blue and impinged by a synthetic promoter, Bba_R0040 (Px, blue arrow), promoterless Gfp gene in light green, and circle junction (donor joint) in brown with the right and left ends intersected by the core CT (blue). The bridge RNA is shown as a dotted line. Bottom: linear depiction of the plasmids and recombinant product. Upper map: Target plasmid with divergent promoters and including the target sequence and transposase gene. Middle map: donor plasmid. Lower map: recombinant plasmid produced by site- (sequence-) specific recombination at the aligned CT dinucleotide cores (blue). Gfp production is driven by the promoter Px and the nc Bridge RNA cannot be expressed because the component which is normally provided by RE is no longer available.
Reprogramming Bridge RNA
The assay was also used to determine whether the target sequences could be changed. A number of changes to the target loop sequence were made (Fig. IS110.25) and tested against wildtype target sequence and the corresponding (complementary) target sequence. The results demonstrated that changes in the ncRNA target loop sequences eliminate integration into the wildtype target sequence but result in robust integration into the corresponding modified target sequences (Fig. IS110.25). This sequence reprogramming provides convincing support for the invasion model (Fig. IS110.23). Although the junction promoter is likely to be strong (that of IS492 is stronger than placuv5; Perkins-Balding. et al 1999 PMID: 10438765), it was also observed that supplying ncRNA in trans from a strong promoter can further increase the activity of ncRNA on integration (in this case for mutant T5, by almost 2 fold).
Target specificity can therefore be modified by changes in the sequence of the target binding loop sequence.
Fig. IS110.25. Integration of Target Loop Variants. The GFP mean fluorescence intensity (MFI) of E. coli after plasmid recombination using the indicated reprogrammed bridge RNA target-binding loop and target sequences (WT and T1–T7). Bold bases highlight differences relative to the WT target sequence. Mean ± s.d. of three biological replicates. None of the target binding loop mutants gave significant activity with a wildtype sequence.
Flexibility in IS621 Target Specificity.
The flexibility of target recognition was further explored [26] using a plasmid-based high throughput method. One plasmid carried the target (Fig. IS110.26A) (together with a promoter), the bridge RNA orf (with the wildtype donor binding loop, DBL) separated by a 12 bp barcode, a chloramphenicol resistance gene and the tnpIS621 gene driven by an inducible T7 promoter (Fig. IS110.26B). The donor plasmid carried the wildtype LE-RE junction (Fig. IS110.26A) (together with an Ampicillin resistance gene and a promoter-less Kanamycin resistance gene). Integration of the donor into the target would bring the inactive kanamycin resistance gene under control of the promoter from the target site and result in KmR recombinants (Fig. IS110.26B).
Fig. IS110.26. Screening for Variation in Target Site Sequence Recognition. Top: A) The screen used a library of variable target (Rep) sequences (shown by the red N nucleotides, top left) and a wildtype donor sequence (bottom left) together with a library of bridge RNAs with a library (right) of variable TBL sequences (red N nucleotides, top right) and a wildtype DBL (bottom right). The blue boxes of the donor and target sequences indicate the complementary strand to those in the TLB and DLB sequences. B) the target plasmid including the barcode, symbols are the same as those shown in Fig. IS110.23. Integration results in activation of the KmR gene.
The target and TBL were cloned as a single oligonucleotide (Fig. IS110.27). The core CT dinucleotide was retained in all cases. Non-CT (core) target and corresponding LTG and RTG positions were then varied to assess single and double mismatch tolerance at each position. For this, several oligonucleotide sets were used and cloned by the Gibson method into a vector plasmid carrying the downstream donor binding loop (Fig. IS110.27). These were designed to test: 1) different target guides with single mismatch pairs; 2) double TBL and target mismatches; 3) negative controls ensuring none of the 9 programmable positions (excluding the CT core) matched in the TBL and target; 4) additional single mismatch combinations in TBL and target; 5) how mismatches in the dinucleotide CT core of the bridge RNA sequences affected recombination efficiency.
The results demonstrated that: full complementarity between the target and TBL was highly preferred (both single and double base mismatches severely impacted integration); integration occurred with sequence complementary changes over all positions in the target and TBL could be reprogrammed and reprogramming showed a large degree of flexibility over all positions.
Fig. IS110.27. Cloning of the Oligonucleotide Library. The plasmid used to clone the oligo nucleotide includes the wildtype DBL, a pT7-driven transposase gene and a CmR gene. The oligonucleotide insert contains the mutant target site, two synthetic and divergent promoters, Bba_R0040 (used to drive the KmR gene in the recombinant product) and a J23119 consensus promoter (used to drive expression of the recomposed nc Bridge RNA) separated by the 12 bp barcode sequence and followed by the TBL mutant sequence.
Insertion in vivo: Reprograming the Target site.
In vivo insertion into the E. coli genome was investigated using a conditional replication defective plasmid with a 22bp wildtype IS621 donor sequence and a wildtype IS621 bridge RNA. Following inhibition of plasmid replication while maintaining selection of a plasmid selective marker, 144/173 unique insertions were identified in known Rep sequences: 96% occurred in the naturally observed target sequence (ATCAGGCCTAC) with only 2 with the exact target binding loop sequence (ATCGGGCCTAC) suggesting that the mismatch which would create an rG:dT base pair might be important; 4/10 of the most frequent integration sites may use an extended base-pairing of RTG and RT (i.e. 7 instead of 4 bp) since they are flanked by 5’-GCA-3’ which is complementary to the 5’-UGC-3’ immediately 5’ the RTG (red ribonucleotides in Fig. IS110.22A). Indeed, many of the orthologues naturally include longer RTGs (purple lozenges in Fig. IS110.22A).
Two reprogrammed bridge RNAs were designed to target two unique E. coli target sequences each with a 4 or a 7 RTG/RT base-pairing. While the most frequent insertion sites were observed to be those expected, some off-site insertions were also observed. These were greatly reduced with the extended 7 nt RTG compared to the 4 RTG bridge RNAs.
Reprograming the Donor site
The fact that the IS621 donor sequence was observed to be more conserved than the target sequence (Fig. IS110.22B) may render it more difficult to reprogram. To examine this, a system similar to that used in reprograming the target site was used but in which the bridge RNA was produced in cis from the donor junction sequence (Fig. IS110.28). Recombination was, again, designed to activate a KmR gene. Similar to the results of target-TBL sequence variation, donor- DBL mismatches significantly reduced activity.
Fig. IS110.28. Screening for Variation in Donor Site Sequence Recognition. A) The screen used a library of variable donor (LE-RE junction) sequences (shown by the red N nucleotides, bottom left) and a wildtype donor sequence (top left) together with a library of bridge RNAs (right) with variable TBL sequences (red N nucleotides, bottom right) and a wildtype TBL (top right). The blue boxes of the donor and target sequences indicate the complementary strand to those in the TLB and DLB sequences. B) the target plasmid. Integration results in activation of the KmR gene.
Insertion in vivo: Reprograming the Donor site
The insertion activity of donor sequences was determined with the Gfp assay used to examine the target sequences. A number of donor mutants and their paired DBL (Fig. IS110. 29: 1-9) were combined with a target sequence (Fig. IS110.25: 5) and its paired TBL sequence. The reprogrammed donor bridge RNAs yielded between 27 and 95 % of wildtype activity (Fig.IS110.29) whereas the wt donor performed poorly with each of the mutants. The reaction was dependent on an intact RuvC domain in the transposase.
This confirmed that, like the target loop, the donor loop sequences can be reprogrammed.
Fig. IS110.29. Integration of Donor Loop Variants. The GFP mean fluorescence intensity (MFI) of E. coli after plasmid recombination using the indicated reprogrammed bridge RNA donor-binding loop and donor sequences (WT and 1–9). Bold bases highlight differences relative to the WT donor sequence. Mean ± s.d. of three biological replicates was included in the original figure.
Analysis of a Second IS110 Group Member: ISEc21.
In addition to IS621, results of a detailed study of another IS110 group member, ISEc21 have shown that an RNA from the upstream NCR region is involved in interaction with the ISEc21 target DNA [25]. ISEc21 was identified in 5 copies in the E. coli E2348/69 chromosome each with an identical target sequence (Iguchi and Hayashi, 2008. Direct submission to ISfinder). The target sequence was confirmed by Siddiquee et al., [25] and, furthermore, shown to be a sequence including and surrounding the central D of the DDE motif of IS3 family members (e.g. ISCfr6, ISEc92, ISEc93).
ISEc21 Transposition: Circle Formation, a Requirement for the NCR and the Target Site
The requirements for transposition activity were examined using a plasmid-cloned ISEc11 copy including ~100bp of flanking DNA (Fig. IS110 30 top). Abutted IS ends, presumably circular transposition intermediates, were detected by PCR, and the junction sequence with the junction promoter determined (Fig. IS110.30 top). Deletion of the upstream NCR sequence (bp 20 – 150) eliminated detectable circles. In addition, insertion into a suitable target DNA (involving both circle formation and insertion) was monitored by PCR reactions at both insert juntions (Fig. IS110.30 A) and was eliminated by deletion of the NCR (Fig. IS110.30 B). However, providing NCR in trans under control of a T7 promoter on a third plasmid, restored the entire reaction (Fig. IS110.30 C).
This system was also used to investigate the target sequence requirements (Fig. IS110.31) which, although not systematic, clearly demonstrated that target specificity was robust and depended on a surprisingly small number of conserved nucleotides: 5/6 consensus nucleotides on the left and 5 on the right or only 3 on the right still permitted IS circle formation and insertion (Fig. IS110. 31).
Fig. IS110.30. The ISEc21 Transposition System. : Donor plasmid (grey circle); transposase gene, tnpEC21 (lilac); upstream non-coding region, NCR, left and right ends, LE and RE, (yellow); flanking sequences (green); ampicillin resistance gene (red). Junction formation was monitored by PCR Top: excision of the IS circle from the donor plasmid. Below: DNA sequence of the circle junction -10 and -35 junction promoter components (grey boxes) ; the left, LE, and right, RE, ends (yellow boxes). A, B and C) target plasmid backbone (red circle). Kanamycin resistance gene (red). ISEc21-target junction formation (insertion) was monitored by PCR at both ends. A) Insertion assay with a wildtype ISEc21. B) ISEc21 without its upstream NCR. C) NCR supplied in trans [25].
Fig. IS110.31. Defining the ISEc21 Target Sequence. Top: Sequence of the target and DNA flanks at the left and right IS ends. Left (LE) and right (RE) IS ends are in yellow boxes. Sequence of PCR products containing the Left flank, LF/LE, and right flank, RE/RF, junctions compared to the target. Identity of the target (green) sequence with the LE and RE flanks is represented by “:”. Bottom: Essential Base pairs in the Target for Integration. Various “target” sequences are shown. The insertion point is indicated by a yellow box. Conserved target bases (green, upper case); adjacent bases and bases altered in the target (black lowercase). Detection of LF/LE and RF/RE junctions by PCR is shown by + or – on the right [25].
Involvement of NCR RNA
Small RNA was recovered associated with TnpISEc21 during purification. RNA seq. of this material produced a strong but extended peak in the upstream NCR (Fig. IS110.32A a). This was of three principal lengths which mapped to the upstream NCR region: nt 1-281, 90-163 and 90-147 (Fig. IS110.32A b). The position of the 3 sRNA spans a region which includes identities to the left and right halves of the of the target site while the entire ISEc21 NCR region, if expressed in its enrirety would also span sequences with identity to the donor site (Fig. IS110.32A c) as has been found by Durrant et al [26] for IS621. The reason for this difference is unclear but in view of the results from their studies on IS1111 group members (in particular ISPa11; Fig. IS110. 35B), it seems probable that the longer RNA is biologically relevant and, we find, carries both the target guides and the downstream donor guides (not shown). Siddiquee et al., [25] have called this sRNA seek RNA since it shows complementarity to the target.
The activities of these sRNA in an in vivo coupled reaction involving excision and insertion of a derivative IS circle were tested in a system in which insertion could be monitored by activation of an mCherry gene (Fig. IS110.32B). All constructs except RNA 90-163 gave positive results in this assay (Fig. IS110.32A b). One explanation for the absence of activity of this RNA is that the region between nt 147 and 163 may generate a structure unable to pair with the target sequence.
Fig. IS110.32. A) Organization of ISEc21. a) Map. ISEc21 (yellow horizontal box) with scale in base pairs above; transposase gene (lilac box) and direction of expression (arrowhead); NCR falls within the blue brackets Above shows the results of RNA seq (red) with coordinates in bp indicated. b) Expanded map showing the sRNA species identified (blue) and their capacity to facilitate integration in the mCherry assay (Fig. IS110.32 B). Dotted lines are linked to the sequence of the NCR and show the left target guide (LTG) and the right target guide (RTG) sequences (green in grey boxes). Also shown are potential right donor guide (RDG) and left donor guide (LDG) sequences (organge in grey boxes). The yellow boxes represent the sequences of the LE-RE containing circle junction. c) Predicted secondary structure of the sRNA showing the position of the LTG and RTG (green) B) mCherry Transposition Assay. Donor plasmid with a promoter-less mCherry gene (pink) flanked by LE and RE (yellow) in turn, flanked by the left and right halves of a target sequence (green); the donor also contains a transposon gene (lilac) and the cloned RNA containing ISEc21 segment (yellow) with a downstream HDV ribozyme (orange) and transcription terminator (blue). Expression is driven by a phage T7 promoter. The target plasmid (red circle) carries a target sequence (green) and a proximal T7 promoter together with a kanamycin resistance gene (red). Excision of the mCherry circle from the donor as a consequence of transposase and NCR expression and its insertion into the target plasmid should result in mCherry expression (deep pink).
Exploring Bridge RNA Secondary Structures from Other IS110 Group Members
Durrant et al [26] also undertook a short survey to determine whether other members of this family also exhibited an RNA with similar structure to the IS621 bridge RNA. A bridge RNA was predicted in nearly 86% of IS110 group members in their library using the RNA covariance models. These were largely located at the left end (see also Fig. IS110.6A). Three IS potential bridge RNAs were examined for complementarity to their donor and target sites. These are shown in Fig. IS110.33.1-3 and their position on the phylogenetic tree is shown in Fig. IS110.3A. Perhaps surprisingly they include a diverse collection of secondary structures.
Fig. IS110. 33. Predicted Bridge RNA from 3 IS110 group Members. Top of the figure shows a map of the IS as a yellow horizontal box containing a purple arrow representing the transposase gene and its direction of expression. The predicted secondary structure is shown below within the blue dotted line which also indicates its location on the IS, its polarity (5’ and 3’ ends), the IS name and length in nucleotides. A code showing the meaning of the symbols is included on the right. The structure shows the left and right target guide sequences (LTG and RTG) as green ellipses and the left and right donor sequences (which interact with the RE-LE junction; LDG and RDG) as brown ellipses. These interactions are indicated in the box on the right with the target and donor sequences appropriately color coded. For ISPpu10 and ISAar29, the bridge RNA sequence is included below with LTG, RTG, LDT and RDT sequences appropriately color coded in bold underlined and boxed and the blue lines indicating the orientation. The interaction of these sequences with the target and donor DNA is presented at the bottom with the complementary sequences boxed. The yellow boxes on the left represent the left and right ends of the IS in the donor junction joint, the substrate which undergoes integration.
RNA from IS1111 Group Members.
Following the proposal that IS1111 group members might use an RNA in the downstream NCR for targeting and integration [29] (Fig. IS110.19), the Hall group chose the IS1111 group member ISEc11 as a model but also investigated other IS1111 members, ISKpn4, ISPst6 and ISPs25 (which all target one end of certain attC integron cassette sites, ISPa11 which targets REP sequences), ISXne4, and an IS110 member (ISEc21; see above). Their positions in the phylogenetic tree is shown in Fig. IS110.3A.
ISEc11, A Model IS1111 Group Member and Some Others.
ISEc11 (Fig. IS110.34) was isolated originally from an enteroinvasive E. coli (EIEC) strain and is located both on the chromosome and on a large (260-kb) F-like virulence plasmid (pINV) (Prosseda et al 2009 PMID: 16788177). Southern hybridization showed that it was present in 9 EIEC strains with differences in the number and the relative location of the chromosomal copies: five East African EIEC strains carry 4 ISEc11 copies in the same position, while the in the remaining four the number varies from 0 to 4. Abutted IS ends, presumably circular transposition intermediates, were detected by PCR. They shared a potential target target sequence, 5’-GTNAAAANANTG-3’, and were all inserted in the same orientation. It was proposed that insertion generated a 4bp DR (5’-AAAT-3’).
Functional Analysis
Using a system similar to that used in analysing ISEc21 (Fig. IS110.30) with a target plasmid into which a specific target sequence is inserted and a donor plasmid carrying either a full ISEc11 copy (Fig. IS110.30A), a copy deleted for the NCR (NCR; Fig. IS110.30B), or a with an additional plasmid which provides the NCR expressed in trans (Fig. IS110.30C), it was demonstrated that the downstream NCR was necessary for transposition and could be supplied in trans from another plasmid. Moreover, in the sequence of the circle junction Prosseda et al (2009 PMID: 16788177) proposed a 4bp target DR. This has now been included within LE where it would contribute to the -10 promoter component. PCR was used to identify the IS circle junction (Fig. IS110.34d) and determine its sequence, revealing the formation of the probable junction promoter. Definition of the target sequence and its use in the target plasmid (Fig. IS110.30) confirmed the expected ISEc11 LE and RE flanks in the insertion products (Fig. IS110.34e) while mutation of the flanking sequences (Fig. IS110.34f) inhibited both circle formation and integration.
Fig. IS110.34. A) Organization of ISEc11. a) Map. ISEc11 (yellow horizontal box) with scale in base pairs above; transposase gene (lilac box) and direction of expression (arrowhead); NCR falls within the blue brackets. Above shows the results of RNA seq (red) with coordinates in bp indicated. b) Expanded map showing the NCR RNA sequence with the left target guide (LTG) and the right target guide (RTG) sequences (green in grey boxes) and their location on the target sequence below. c) Predicted secondary structure of the sRNA showing the position of the LTG and RTG (green). d) IS circle junction. LE and RE (yellow boxes); -10 and -35 promoter components (grey boxes); Subterminal inverted repeats (red text within grey arrows). e) Sequence of the target and DNA flanks at the left and right IS ends. Left (LE) and right (RE) IS ends are in yellow boxes. Sequence of PCR products containing the Left flank, LF/LE, and right flank, RE/RF, junctions compared to the target. Identity of the target (green) sequence with the LE and RE flanks is represented by “:”. f) Transposition with altered target sequences flanking ISEc11 and in pTarget. (see Fig. IS110.30 for reference) Sequences tested are on the left with consensus target bases green and the boundaries between IS and target indicated by a yellow box [25].
Identification of IS1111 Group ncrRNA
Like that of IS621, an RNA, ncrRNA, was found to copurify with the ISEc11 transposase and its presence increased transposase yield. RNA seq revealed a peak located within the NCR located downstream of the transposase, tnpEc11, gene (Fig. IS110.34a). This yielded two principal species of ~80 and 150 nt (82-164 and 82-227; Fig. IS110.34a) although the RNA peak was somewhat disperse. Similar results identifying a long and shorter sRNA were obtained with 5 additional IS1111 group members ISKpn4 (Fig. IS110.35A), ISPa11 (Fig. IS110.35B), ISPst6 (Fig. IS110.35D), ISPa25 (Fig. IS110.35E) and ISXne4 (Fig. IS110.35F). While ISPst6 is very similar to ISKpn4 (Fig. IS110.35Dc and 35Ec), has identical IRst sequences and a Tnp 86% identical and 92% similar to TnpISKpn4, ISPa25 is more distant: TnpISPa25 and TnpISKpn4 and are 46% identical and 60% similar (Fig. IS110.35Ec). ISKpn4, ISPst6 and ISPa25 fall into the same IS clade (Fig. IS110.3A) and Interestingly the RTG and LTG are nearly identical and identically spaced (Fig. IS110.35Ed) reflecting their similar target sites.
Fig. IS110.35. A) Organization of ISKpn4. a) Map. ISKpn4 (yellow horizontal box) with scale in base pairs above; transposase gene (lilac box) and direction of expression (arrowhead); NCR falls within the blue brackets. Above shows the results of RNA seq (red) with coordinates in bp from the tnp stop codon indicated. b) Expanded map showing the NCR RNA sequence with the left target guide (LTG) and the right target guide (RTG) sequences (green in grey boxes) and their location on the target sequence below. c) Predicted secondary structure of the sRNA showing the position of the LTG and RTG (green) B) Organization of ISPa11. Features are indicated as in A). a) Map. b) Expanded map showing the NCR RNA sequence with the left target guide (LTG) and the right target guide (RTG) sequences (green in grey boxes) and their location on the target sequence below. Also shown are potential right donor guide (RDG) and left donor guide (LDG) sequences (orange in grey boxes). The yellow boxes represent the sequences of the LE-RE containing circle junction. c) Predicted secondary structure of the sRNA showing the position of the LTG and RTG (green). C). Predicted LTG/RTG and LDG/RDG in the downstream ISPa11 NCR from Durrant et al [26]. D) Organization of ISPst6. Features are indicated as in A). a) Map. b) Expanded map with the NCR RNA sequence and left (LTG) and right (RTG) target guide sequences (green in grey boxes) and their location on the target sequence below. c) Alignment of ISKpn4 with ISPst6. E) ISPa25 a) Map b) Expanded map with the NCR RNA sequence and left (LTG) and right (RTG) target guide sequences (green in grey boxes) and their location on the target sequence below. c) Alignment of ISPa25 and ISPst6 on ISKpn4. Identities are shown in red. d) Alignment of RTG and LTG of ISKpn4, ISPst6 and ISPa25 [25].
Additionally, Siddiquee et al., [25] identified the equivalent of LTG and RTG in the smaller, majority, RNA from all five IS1111 group IS (Fig. IS110.34; Fig. IS110.35A and 35B), but the short RNA sequence did not include the donor LDG and RDG sequences. It was noted that the order of LTG and RTG within the IS1111 IS NCR RNA was inverted compared to that found for the IS110 group, ISEc21 (Fig. IS110.32Ab), an observation also made by Durrant et al [26]; Fig. IS110.35C; Fig. IS110.36A and 36B). Since the short RNA would have affinity for the target site but not the donor site, it was called RNA seek. However, the longer RNA (not shown) also includes sequences resembling LTD and RTD.
This is illustrated in the case of ISPa11 analysed by both Siddiquee et al., [25] and Durrant et al [26] but can also be seen in the other IS. Inspection of the short RNA sequence of Siddiquee et al (Fig. IS110. 35Bb) shows that it terminates within a potential LDG signal. Extending this RNA sequence uncovers not only an LDG but a corresponding RDG which would be present in the long RNA species (Fig. IS110. 35Bb). Again, the LDG and RDG are inverted with respect to the IS110 group members. These sequences were those predicted by Durrant et al [26] (Fig. IS110. 35C). A similar arrangement was also exhibited by two additional IS1111 group members ISCARN28 and ISAzs32 [26]; Fig. IS110.36A and 36B).
Other IS1111 Group Members.
As in the case of the IS110 group, Durrant et al [26] also undertook a short survey of members of the IS1111 group to identify RNA with similar structure to the IS621 bridge RNA. In addition to those shown in Fig. IS110.35C and 36, a bridge RNA was predicted in 93% of IS1111 group members in the library using the RNA covariance models. These were largely located in the right end (see also Fig. IS110.6A).
Fig. IS110. 36. Predicted Bridge RNA from 3 IS1111 group Members. Top of the figure shows a map of the IS as a yellow horizontal box containing a purple arrow representing the transposase gene and its direction of expression. The predicted secondary structure is shown below within the blue dotted line which also indicates its location on the IS, its polarity (5’ and 3’ ends), the IS name and length in nucleotides. A code showing the meaning of the symbols is included on the right. The structure shows the left and right target guide sequences (LTG and RTG) as green ellipses and the left and right donor sequences (which interact with the RE-LE junction; LDG and RDG) as brown ellipses. These interactions are indicated in the box on the right with the target and donor sequences appropriately color coded.
Programming ISEc11 Integration.
Siddiquee et al.,[25] tested whether, like the IS110 member Bridge RNAs (Fig. IS110.25 Fig. IS11029; [26], the IS1111 group Seek RNA can be reprogrammed to recognize both alternative target sites. This was explored using ISEc11 in the mCherry assay system (Fig. IS110.32B). Transposition was measured by flow cytometry as the percentage of mCherry expressing cells in the population. Two modified long seek RNAs together with the corresponding modified LE and RE flank sequences in the donor gave robust transposition (Fig. IS110.37e and f) although their target activities were not tested with wildtype seek RNA. It is interesting to note that the short wildtype seek RNA was significantly more efficient in promoting transposition than the long wildtype seek RNA (compare Fig. IS110.37c and d).
Fig. IS110.37. Reprogramming seekRNA. Both the LE and RE flanks and the target DNA sequences were changed concomitantly. The ISEc11 seekRNA used in the donor plasmid was the long (154 nt) species. Insertion resulted in expression of the mCherry gene carried within two ISEc11 ends from a resident T7 promoter located in the target plasmid (Fig. IS110.32B). The percentage of mCherry-expressing cells in the population was measured by flow cytrometry. c) transposition with wildtype target and long seekRNA, 15% When the portion of the target that flanks the IS on the right was altered and the corresponding changes were made in the seekRNA. d) transposition with wildtype target and short seekRNA, 42% e) transposition to the M1 target occurred at about 23% frequency. f) transposition to the M2 target was 15%.
Use in Genome Modification
Clearly, the use of the mCherry system demonstrates that the IS110 family is capable of delivering a genetic cargo and that TnpISEc11 can be supplied in trans. Siddiquee et al., [25] extended these observations to demonstrate that the ~750bp chloramphenicol acetyltransferase gene (CAT) can also be inserted either upstream or downstream of the tnpISEc11 gene and that the ISEc11 derivative remains transpositionally active. Additionally, Durrant et al [26] designed a GFP reporter system for the IS110 member IS621 which allowed them to demonstrate the capacity of this system to generate deletion and inversion events when donor and target are located on the same DNA molecule. The system was designed such that recombination brought the GFP gene under control of a neighboring adjacent promoter. As might be expected from other systems, such as transposon Tn3 family resolution, deletion occurs when the target and donor sites are present in the same orientation where inversion occurs when they are inverted with respect to one another.
Structural Analysis: the Synaptic Complex Involved in Circle integration
Cryo-EM was used to explored the IS621 insertion mechanism in detail [33]. It revealed the organization of the IS621 synaptic integration complex in three different stages of the recombination pathway involved in IS insertion. The complex was assembled using full length (177nt) purified bridge RNA (b-RNA) obtained by in vitro transcription from a T7 promoter (see Fig. IS110.22A), the double stranded RE-LE IS circle junction DNA (j-DNA or d-DNA; 44bp), the double stranded target DNA (t-DNA; 38bp) and purified transposase, TnpIS621, obtained using a standard expression vector. This complex was unstable but could be stabilised by introducing 6 consecutive mismatches in the top strands of d-DNA and t-DNA (positions 2–7; Fig. IS110.38A, top) in TBL and DBL. The structure was solved at 2.5 Å resolution.
It was composed of: 4 TnpIS621 monomers (A-D) (Fig. IS110.38A, bottom left), both TBL and DBL segments of the b-RNA and both t- and d-DNA. The 5’ b-RNA stem loop (Fig. IS110.24) was not visible, suggesting flexibility, its deletion reduced complex stability implying that it may enhance b-RNA/TnpIS621 Interactions. It was also suggested that that two different b-RNA molecules may contribute the TBL and DBL, respectively.
Fig. IS110.38A IS621 Synaptic Integration Complex (PDB ID:8WT6). Top: t-DNA and d-DNA sequences. left (LTG) and right target guide (RTG) sequences (green in grey boxes). Right (RDG) and left donor guide (LDG) sequences (orange in grey boxes). The yellow boxes represent the sequences of the LE-RE containing circle junction. Blue letters show the core nucleotides. Lower case bold characters indicate the mismatches introduced into the sequences which lead to formation of stable complexes. Below left: synaptic complex. All 4 TnpIS621 monomers are color-coded as are the b-RNA, d-DNA and t-DNA molecules. Below right: configuration of DNA and RNA in the synaptic complex.
Fig. IS110.38B IS621 Synaptic Integration Complex (PDB ID:8WT6). Top: Structure of nucleic acids. The positions of the target (green, left) and donor (orange, right) base pairing with the bridge RNA are circled and enlarged (boxes) [26]. Middle: Schematic of the pairing model. Bottom: Simplified Cartoon of the RNA/DNA structures [33]. Bridge RNA is shown in dark blue, target DNA in green and donor DNA in brown. Left and right target and donor DNA is indicated (LT, RT, LD and RD respectively) as are the left and right Target and Donor and Donor guide sequences (LTG, RTG, LGD and RGD respectively). The active site serine 241 is shown as a yellow circle.
In addition to revealing a composite active site which positions the catalytic serine (Tnp) residues adjacent to the recombination sites in both target and donor DNA. Comparison of the three structures identified showed: strand cleavage of target and donor DNA at the composite active sites to generate 5′-phosphoserine covalent intermediates as found in other recombination systems such as Tn3 family transposon resolution and IS607 transposition; creation of a Holliday junction intermediate by strand exchange and rejoining using a 3’OH generated resulting from formation of the 5′-phosphoserine covalent intermediates; and resolution by second strand cleavage
Synaptic Complex Assembly
The synaptic complex is assembled from the two dimeric TnpIS621 complexes: monomers A and B form a dimer which interacts with TBL and t-DNA while C and D constitute a dimer which interacts with the DBL and d-DNA (shown schematically in Fig. IS110.39). The two dimers contact each other via their RuvC domains. The TnpIS621 monomer is folded into three domains (Fig. IS110.40 right): a coiled-coil domain, CC, containing two α-helices; a “transposase” domain, Tnp, including the active site serine 241; and a RuvC domain carrying the DEDD motif. Protomer dimerization between TnpIS621.A and TnpIS621.B and between TnpIS621.C and TnpIS621.D is mediated by the CC domain (Fig. IS110.40 left). Similar protein structural models were predicted for both IS110 (TnpISEc21) and IS1111 (TnpISEc11 ) family members [25] using AlphaFold. As might be expected, TBL and t-DNA and DBL and d-DNA are base paired (Fig. IS110.38A, bottom right; Fig. IS110.38B; Fig. IS110.39) and t- and b-DNA are bent into an X configuration. Both t- and d-DNA are cleaved bordering the CT core sequences (C8–T9; Fig. IS110.38B, Fig. IS110.39) using the conserved serine (S241; Fig. IS110.8) as the nucleophile and forming a covalent 5’-phosphoserine bond with T10 (Fig. IS110.39). Extra-helical bases A43 and A67 in TBL and A116 and A150 in DBL together with syn conformation G nucleotides G48 and G72 in TBL and G121 and G155 in DBL (Fig. IS110.39 middle and left) are highly conserved in IS110 family members and are recognized in the same way by the Tnp domain by all 4 TnpIS621 monomers.
Opening of the t- and d-DNA Duplexes
The structure also explains how the t- and d-DNA duplexes are destabilized to facilitate their recognition by b-RNA: clustered tyrosine and methionine residues within the Tnp domains wedge between a number of complementary nucleotides in both duplexes (Fig. IS110. 39 middle) and mutation of these amino acids reduces recombination significantly.
Fig. IS110.39. Bridge RNA Interaction with Donor and Target. Bridge RNA is shown in dark blue, target DNA in green and donor DNA in brown. Left and right target and donor DNA is indicated (LT, RT, LD and RD respectively) as are the left and right Target and Donor and Donor guide sequences (LTG, RTG, LGD and RGD respectively). The active site serine 241 is shown as a yellow circle and labelled in a colored box according to the associated Tnp monomer. Left: Model from Durrant et al [26]. The core dinucleotides are within a box. Middle: Simplified Cartoon of the RNA/DNA structures [33]. Extra helical A and syn conformation G nucleotides are shown within blue elipses and their approximate positions indicated by red arrows. The approximate positions of the “wedge” amino acids (Y264, M265 and M268) are shown within colored elipses correspond to each associated monomer. Right: schematic of nucleic acid interactions observed in the structure. Red letters circled in blue indicate conserved extra-helical A and syn configured G. The boxed cartoon illustrated hydrogen bonding between the target and donor sequences.
Fig. IS110.40. TnpIS621 and the Synaptic Complex (PDB ID:8WT7). Right: Structure of monomer D. The structure shows three principal domains: the Tnp domain (yellow circle) showing the position of the catalytic serine 241; the RuvC domain (blue circle) showing the position of D11,102 and 105; and the coiled-coil domain composed of two a-helices. Left: Arrangement of the tetramer. The nucleic acids have been removed. Each monomer in the dimer of dimers is indicated. The figure shows the formation of A/B and C/D dimers via interaction of their coiled-coil domains (CC) and the hybrid or composite A/D and B/C catalytic centers within yellow circles. The acidic residues are shown as red dots and the catalytic serine as a small yellow circle.
Composite Active Sites.
The TnpIS621.B and TnpIS621.D loops carrying S241 interact with those carrying D102 (Fig. IS110.40 right) in TnpIS621.C and TnpIS621.A to form a composite active site between the A/B and the C/D dimer (Fig. IS110.41 left). On the other hand, the S241 loops of TnpIS621.A and TnpIS621.C are disordered and the TnpIS621.B and TnpIS621.D D102 loops have a different conformation to those in TnpIS621.A and TnpIS621.C which form part of the active site.
The TnpIS621 RuvC domain is therefore unusual since it does not act independently, as do other RuvC domains (e.g. IS200/IS605 family TnpB), but functions together with the Tnp domain (i.e. S241) in the composite active site. It was suggested that this arrangement may prevent adventitious DNA cleavage occurring before synaptic complex assembly, a characteristic of a number of other systems such as phage Mu (e.g. Williams et al., 1999 PMID: 10541558) and Tn5/IS50 (Protein structure and the transpososome; Naumann and Reznikoff 2000 PMID: 10908658 ). The RuvC domains also play a central role in synaptic complex formation since the two dimers contact each other through RuvC–RuvC interactions.
Fig. IS110.41 Recombination Steps in Integration. Target (green); Donor (orange); bridge RNA(blue); mismatched bases (lowercase); S241 (yellow circle) with accompanying colored box indicating which monomer is involved; cleavage point (red triangle); co-ordinates from 1-14 are shown. The “Handshake” are indicated by a red box. bases are indicated Left: b-RNA interaction with target DNA. Top and Bottom: t-DNA and d-DNA sequences. left (LTG) and right target guide (RTG) sequences (green in grey boxes). Right (RDG) and left donor guide (LDG) sequences (orange in grey boxes). The yellow boxes represent the sequences of the LE-RE containing circle junction. Blue letters show the core nucleotides. Lower case bold characters indicate the mismatches introduced into the sequences which lead to formation of stable complexes. Middle top: Target DNA and target loop RNA Interaction. Middle bottom: Donor DNA and donor loop RNA. First Strand Cleavage
“Handshaking”
This synaptic complex is, however, trapped in the prestrand-transfer step because of the mismatched base pairs in both t-DNA and d-DNA introduced to stabilize the complex (Fig. IS110.38A top; see also Fig.IS110.22A).
Close examination of the covariation signals obtained with a large number of IS621-related IS (e.g. Fig. IS110.22A) revealed weak additional signals which implied base-pairing potential of nt 6 and 7 of target DNA with the long-distant donor RDG (nt 166) and of nt 6 and 7 of donor DNA with the long-distant donor RTG (nt 81). This was called Handshake base pairing and the sequences were named Handshake guides (HSG). It was noted that they play a role in the first strand exchange reaction. Exchange in the wildtype situation increases the potential base pairing (Fig. IS110.41 Fig.IS110.42 A). Measurement of full recombinants in vitro with wildtype b-RNA (Fig. IS110.42A) showed that in addition to robust recombination products, a significant proportion of cleavage products of the t- and d-DNA had occurred. A series of experiments were designed to examine the effects of Handshake nucleotide complementarity on strand exchange using modified b-RNA. Generating total complementarity of RTG-target and RDG-donor duplex HSG (i.e. prior to strand transfer; Pre-HSG; Fig. IS110.42B) strongly favoured t- and d-DNA cleavage but eliminated detectable recombination in vitro, whereas modifying the HSG sequences to generate perfect complementarity after strand transfer (Post-HSG; Fig. IS110.42C) strongly favored DNA recombination in vitro at the expense of d-DNA cleavage products. The “handshake” dinucleotide therefore clearly strongly influences the outcome of the reaction.
Fig. IS110.42 Modifying Target and Donor Complementarity: The Handshake Dinucleotide. Target (green); Donor (orange); bridge RNA(blue); mismatched bases (lowercase); S241 (yellow circle) with accompanying colored box indicating which monomer is involved; cleavage point (red triangle); co-ordinates from 1-14 are shown; mutated nucleotides are shown in red and new inter-strand bond are shown in red..The “Handshake” are indicated by a red box which, in the case of the strand exchange is extended to include the entire 4 nt that are transfered. A: Wildtype Sequences. Schematics of the TBL/DBL and tDNA/dDNA sequences used for cryo-EM analysis and in vitro recombination assays. B and C: pre- and post-HSB (handshake base-pairing) b-RNAs stabilize the synaptic complex in the pre- and post-strand exchange states, respectively. Mutated nucleotides in the pre- and post-HSB bRNAs and their complementary DNA nucleotides are highlighted [33].
To investigate the steps in the reaction, in addition to the synaptic complex assembled with the 7 mismatches in t- and d-DNA (Fig. IS100.41 left top and bottom; Fig. IS110.43A), structures were resolved using both Pre-HSG b-RNA where recombination is blocked at the pre-strand transfer step (Fig. IS110.42B; Fig. IS110.43B), and Post-HSG b-RNA where recombination is robust but cleavage is reduced (Fig. IS110.42C; Fig. IS110.43C).
Fig. IS110.43A-C. Cryo-EM structure of the IS621 synaptic complex.A) PDB ID:8WT6. Synaptic Complex Stabilised by mismatches in t-and d-DNA.B) PDB ID:8WT7. Pre-HSB b-RNA structure. 1st strands of t- and d-DNA cleaved to form 5′-phosphoserine intermediates. HSGs in TBL and DBL form the expected base pairs with the t-DNA and d-DNA and impede 2nd-strand exchange. C) PDB ID:8WT8. Post-HSB b-RNA.
The cryo-EM structure of the post-HSB b-RNA (Fig. IS110.43C) synaptic complex reveal two states: a post 1st strand exchange trapping the Holliday Junction intermediate and a post strand exchange with HJ resolution. In one (Fig. IS110.44 left) the 1st strand transfer of the donor (at DBL) junction appears complete while that of the target (at TBL) is only partially rejoined while in the other (Fig. IS110.44 right) species, the 2nd strand of the donor (at DBL) junction has been cleaved and the 2nd target strand (at TBL) is only partially cleaved.
Fig. IS110.44 TBL–tDNA and DBL–dDNA post-strand exchange synaptic complexes. Target (green); Donor (orange); bridge RNA(blue); mismatched bases (lowercase); S241 (yellow circle) with accompanying colored boxes indicating which monomer is involved; cleavage (red triangle); partial 1st strand rejoining (left) and partial 2nd strand cleavage (right) (green triangles); red boxes indicate the transferred nucleotides. Left: Holliday junction intermediate state. partial 1st strand rejoining. Right: Holliday junction resolution state. partial 2nd strand cleavage of donor and cleavage of target [33].
These snapshots provide a detailed overall picture of the way in which the IS LE-RE junctions formed to generate circular transposition intermediates interact with their bridge RNAs as the donor DNA and how the bridge RNA interact with the target. Bridge RNA clearly orchestrates the apposition of IS junction and target DNA generating a defined structure
Mechanism Involved in the First Transposition Step: Circle Formation?
However, there are a number of important questions remaining not least, the mechanism by which the IS circular intermediate is generated. Formation using site-specific recombination would be expected to regenerate the original target site. Siddiquee et al., [25] were unable to detect such uninterrupted sequences with the PCR assay used to detect ISEc11 circle intermediates. This suggests that excision does not occur using a classical double-strand site-specific recombination mechanism. It remains possible that excision occurs using a single-strand recombination accompanied by a replicative step in a copy-out-paste-in mechanism similar to that used by the IS3 and other IS families. None of the recent studies have addressed this step of the transposition process.
Long and short: How is IS1111 NCR RNA Generated: Processing?
Another question which arises for both the IS110 and IS1111 groups is how the RNA which co-purifies with the transposase is produced. In the case of ISPa11, no specific NCR promoter was identified by inspection and it was suggested that the small RNA is generated from a longer transcript [25], possibly from the transposase mRNA. This has been demonstrated in the case of the guide RNA from IS200/IS605 family members where the TnpB guide endonuclease is involved (IS200/IS605 family: RNA Nomenclature, Processing, Structure, Diversity and mode of function). It probably also occurs in generating the upstream RNA virulence repressor of IS200, arc200, from the tnpA mRNA (Fig IS200.74) (Ellis et al 2017 PubMed:28335027). It would be interesting to determine whether the presence of the shorter seek RNA require transposase catalytic activity.
Is there a Biological Significance to the High Level of the shorter Seek RNA species?
The observation that the shorter sRNA species is the major RNA product which purifies with the transposase of both IS1111 group members (ISEc21, ISKpn4 and ISPa11; Fig. IS110. 34, 35A, 35B) and IS110 group member, ISEc21; Fig. IS110.32A) and that the longer RNA is significantly less abundant is intriguing. A trivial explanation would be that it has a higher affinity for the transposase than bridge RNA. The short RNA was not identified by Durrant et al [26] presumably because their approach would not necessarily have detected such species. One notion would be that rather than a degradation product, the small seek RNA is in some way involved in IS circularization for example, by recognizing the two flanking segments of the target sequence. Another possibility is that it acts in trans to “prime” suitable targets in the host genome for recognition by the IS circle.
Additionally, is the long RNA carrying the LDG and RDG sequences required for integration or is it involved in assuring the formation of the IS circle? Do both short and long RNA have similar affinity for the transposase?
Possibility of regulation by arc9-like anti RNA.
An important consideration is the regulatory role and presence of anti-RNA such as ars9 found in ISPpu9 [30] in other IS110 family members. This, to our knowledge, has not received further attention. It should be noted that an upstream NCR (UTR) in the unrelated IS200 (IS200 Regulation and Salmonella Pathogenicity) is processed to become a repressor of transcription of certain Salmonella host virulence-associated genes (Ellis et al. 2017 PMID: 28335027). Expression of an anti-RNA, art200, leads to RNA-anti-RNA interactions between complementary secondary structures in the NTR and degradation of transposase mRNA (including the 5’ processed NCR region). It therefore seems possible that, because of their similar organisation, IS110 family members might also be regulated in this way.
Take Home Messages.
It is important to note that there are some ambiguities in a number of the ends of IS110 family members documented in ISfinder due to the absence of terminal IRs. As pointed out by Siddiquee et al., [25], the most definitive method of resolving these problems would be to obtain the DNA sequence of the RE-LE IS circle junction.
Bibliography
- ↑ <pubmed>2993819</pubmed>
- ↑ 2.0 2.1 <pubmed>16381877</pubmed>
- ↑ <pubmed>PMC206497</pubmed>
- ↑ <pubmed>PMC267840</pubmed>
- ↑ <pubmed>PMC268225</pubmed>
- ↑ <pubmed>9526198</pubmed>
- ↑ <pubmed>1685008</pubmed>
- ↑ <pubmed>PMC228102</pubmed>
- ↑ <pubmed>PMC265507</pubmed>
- ↑ <pubmed>9375297</pubmed>
- ↑ <pubmed>PMC2104537</pubmed>
- ↑ <pubmed>PMC154366</pubmed>
- ↑ <pubmed>22850965</pubmed>
- ↑ <pubmed>PMC219399</pubmed>
- ↑ 15.0 15.1 15.2 15.3 15.4 15.5 15.6 <pubmed>9933934</pubmed>
- ↑ <pubmed>PMC280332</pubmed>
- ↑ <pubmed>PMC209814</pubmed>
- ↑ 18.0 18.1 18.2 18.3 18.4 18.5 18.6 18.7 <pubmed>12897009</pubmed>
- ↑ 19.0 19.1 <pubmed>PMC1112027</pubmed>
- ↑ <pubmed>PMC208434</pubmed>
- ↑ <pubmed>PMC178977</pubmed>
- ↑ 22.0 22.1 22.2 22.3 22.4 22.5 <pubmed>PMC166490</pubmed>
- ↑ <pubmed>PMC205616</pubmed>
- ↑ 24.0 24.1 24.2 24.3 <pubmed>PMC545610</pubmed>
- ↑ 25.00 25.01 25.02 25.03 25.04 25.05 25.06 25.07 25.08 25.09 25.10 25.11 25.12 25.13 25.14 25.15 25.16 25.17 25.18 25.19 25.20 25.21 25.22 <pubmed>38898016</pubmed>
- ↑ 26.00 26.01 26.02 26.03 26.04 26.05 26.06 26.07 26.08 26.09 26.10 26.11 26.12 26.13 26.14 26.15 26.16 26.17 26.18 26.19 26.20 26.21 26.22 26.23 26.24 26.25 26.26 26.27 26.28 26.29 26.30 <pubmed>38328150</pubmed>
- ↑ 27.0 27.1 27.2 27.3 27.4 27.5 27.6 27.7 27.8 27.9 <pubmed>38926615</pubmed>
- ↑ <pubmed>18487340</pubmed>
- ↑ 29.0 29.1 29.2 <pubmed>19025573</pubmed>
- ↑ 30.00 30.01 30.02 30.03 30.04 30.05 30.06 30.07 30.08 30.09 30.10 <pubmed>34379788</pubmed>
- ↑ 31.0 31.1 31.2 31.3 31.4 31.5 <pubmed>14563872</pubmed>
- ↑ <pubmed>11169105</pubmed>
- ↑ 33.0 33.1 33.2 33.3 33.4 33.5 <pubmed>38926616</pubmed>
- ↑ <pubmed>3167979</pubmed>
- ↑ <pubmed>7923356</pubmed>
- ↑ <pubmed>10092658</pubmed>
- ↑ <pubmed>1326596</pubmed>
- ↑ 38.0 38.1 38.2 38.3 <pubmed>PMC93982</pubmed>
- ↑ <pubmed>PMC1794265</pubmed>
- ↑ 40.0 40.1 40.2 40.3 40.4 <pubmed>PMC1483014</pubmed>
- ↑ 41.0 41.1 41.2 41.3 41.4 <pubmed>2575701</pubmed>
- ↑ <pubmed>8065263</pubmed>
- ↑ 43.0 43.1 43.2 43.3 <pubmed>11523772</pubmed>
- ↑ <pubmed>14563872</pubmed>
- ↑ <pubmed>19025573</pubmed>
- ↑ <pubmed>38898016</pubmed>
- ↑ 47.0 47.1 <pubmed>2177525</pubmed>
- ↑ 48.0 48.1 <pubmed>8389980</pubmed>
- ↑ 49.0 49.1 <pubmed>1700062</pubmed>
- ↑ <pubmed>PMC2753022</pubmed>
- ↑ <pubmed>9933934</pubmed>
- ↑ <pubmed>PMC125674</pubmed>
- ↑ <pubmed>PMC1169952</pubmed>
- ↑ <pubmed>11069682</pubmed>
- ↑ <pubmed>10471285</pubmed>
- ↑ <pubmed>26350330</pubmed>
- ↑ 57.0 57.1 <pubmed>PMC1207841</pubmed>
- ↑ 58.0 58.1 <pubmed>8459773</pubmed>
- ↑ 59.0 59.1 <pubmed>8057840</pubmed>
- ↑ <pubmed>10673002</pubmed>
- ↑ <pubmed>PMC2817692</pubmed>
- ↑ <pubmed>PMC3686654</pubmed>
- ↑ 63.0 63.1 <pubmed>PMC1317595</pubmed>
- ↑ <pubmed>PMC113213</pubmed>
- ↑ 65.0 65.1 65.2 65.3 <pubmed>PMC1525189</pubmed>
- ↑ 66.0 66.1 <pubmed>PMC2447020</pubmed>
- ↑ <pubmed>19025573</pubmed>
- ↑ 68.0 68.1 <pubmed>16845431</pubmed>
- ↑ <pubmed>1662753</pubmed>
- ↑ 70.0 70.1 <pubmed>19730680</pubmed>
- ↑ <pubmed>20707672</pubmed>
- ↑ <pubmed>16641988</pubmed>
- ↑ <pubmed>15036538</pubmed>
- ↑ <pubmed>29708644</pubmed>