Difference between revisions of "IS Families/IS1595 family"
| (One intermediate revision by the same user not shown) | |
(No difference)
| |
Latest revision as of 12:51, 5 December 2022
Contents
General
The IS1595 family emerged during an analysis of IS1 family members. Using the clustering software, TribeMCL[1], the IS1 family was found to be linked to another new IS group including IS1595 (Fig.5.1). first identified in Xanthomonas campestris (AF249895) [2]. IS1595 was closely related to other IS (e.g. ISXo2, ISXo5, ISXo16, and ISXca4) present in high copy number in other Xanthomonas species. The IS1595 family is less homogeneous than the IS1 family. BLAST analysis of ISfinder using IS1595 as a query confirmed a distant relationship with IS1 family members and with IS1016 [3], a multicopy Neisseria element previously binned with the “unclassified” IS (See "ISNYC"). Seven subgroups have now been identified: IS1595, IS1016, ISPna2, the halobacterial ISH4 group [4], ISSod1, ISNwi1 and ISNha5.
Members of the ISPna2, ISNwi1 and ISNha5 subgroups may contain passenger genes or additional noncoding DNA [2] (Table IS1 1 and Table IS1 2). Only a single example of each member was identified, suggesting that these IS have low or no transposition activity. It is also related to ISSag10 from Streptococcus agalactiae, originally called MTnSag1 and thought to be a member of the IS1 family[5] but now called tISSag10 due to the presence of a passenger gene, an O-lincosamide nucleotidyltransferase. tISSag10 also carries an origin of transfer and can be mobilized by transfer functions of Tn916. It can therefore also be defined as a non-autonomous ICE, this underlines the increasing difficulty in TE classification (see "Fuzzy Borders" in General Information).
Organization
Most are flanked by 8 bp AT-rich DR and have a single Tpase orf. Like IS1 family Tpases, they include an N-terminal ZF, an HTH motif, and a C-terminal catalytic motif with some exceptions (Fig.IS1.6). For example IS1016 group members lack the N-terminal ZF but, as IS1016 is present in multiple copies, it is probably active (Table IS1 1; Fig.1595.1). Not all members of the ISSod11 and ISNha5 groups have a clearly predictable HTH motif (Table IS1 1; Fig.1595.2; Fig.1595.3), but do show a good alignment in this region.
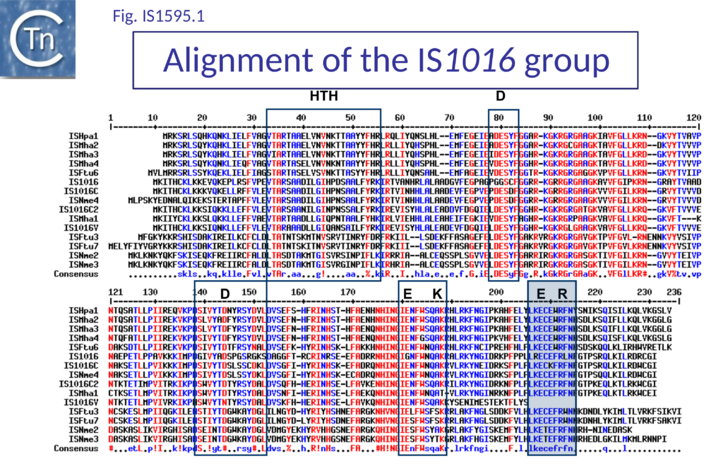
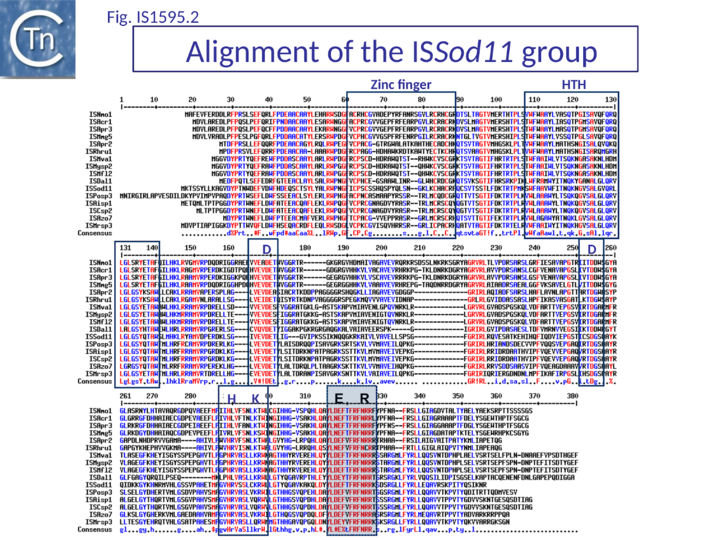
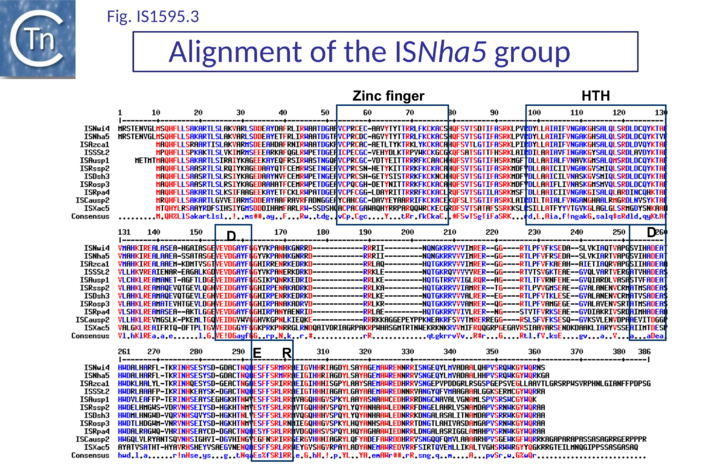
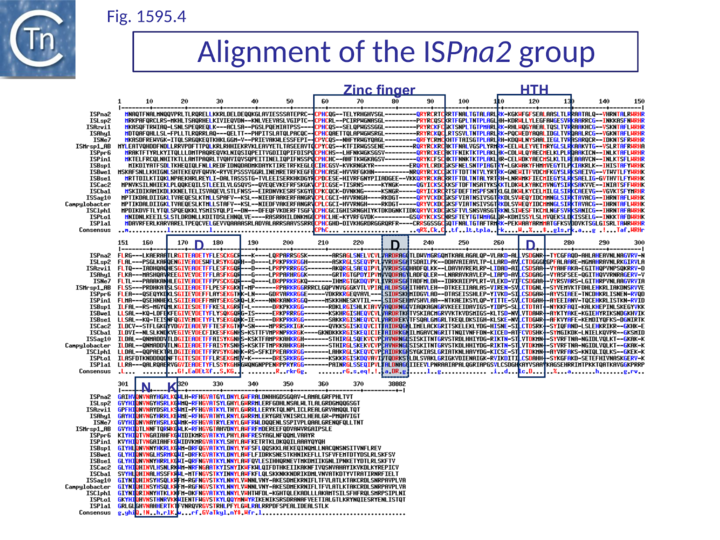
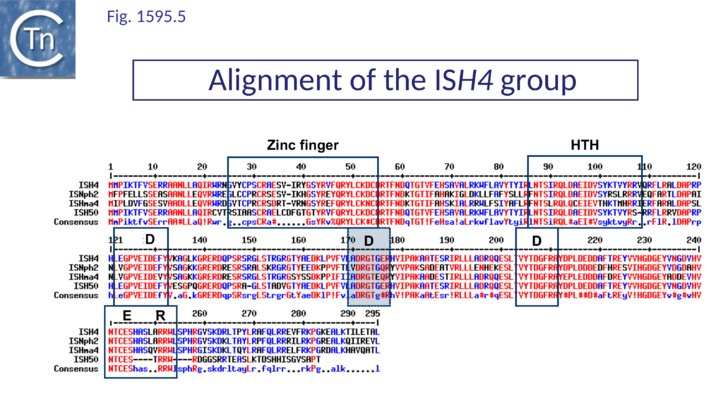
The catalytic sites of all family members show group-specific variation particularly around the final E residue (Fig. IS1.5B).. ISPna2 (Fig.1595.4) and ISH4 (Fig.1595.5) group members include an additional D residue with similar spacing which does not occur at the same position as either of the D residues of the IS1 or other IS1595 family transposases (Fig. IS1.5B)(Fig.1595.5). More surprising is the apparent substitution of the final E residue for N or H in certain members.
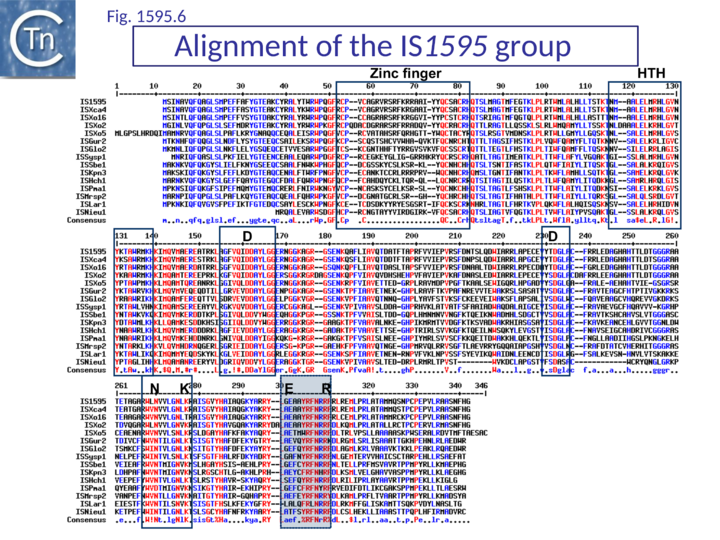
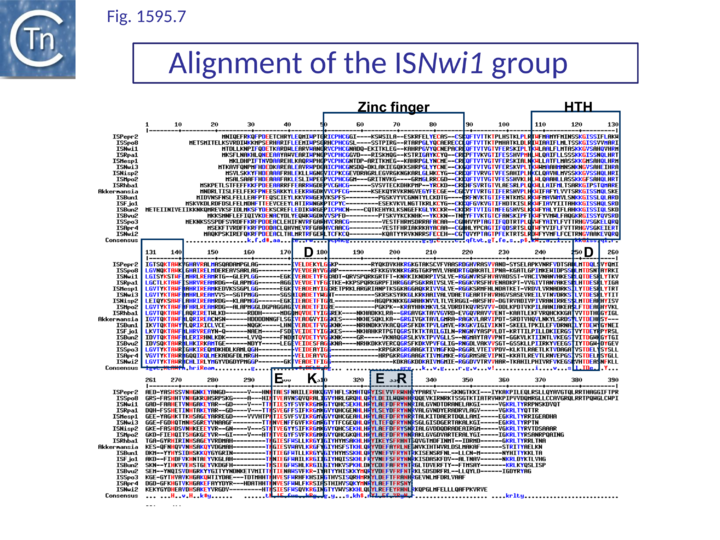
The exact nature of these possible non-canonical catalytic sites will require experimental determination. While ISH4 (Fig.1595.5), IS1016 (Fig.1595.1), ISNwi1 (Fig.1595.7) and ISNha5 (Fig.1595.3) members exhibit a classic DDE7K/R, ISPna2 (Fig.1595.4) and IS1595 (Fig.1595.5) members carry DDN7K at this position, and ISSod11 (Fig.1595.2) carries a DDH7K signature which aligns with the classical conserved DDE7K. Moreover, in four groups (IS1016, IS1595, ISSod11 and ISNwi1), a potential E residue with an associated R lies further downstream but does not align with that of all other members of the family.
It has been reported that the IS1595 family (in particular IS1016) is related to the Merlin family of eukaryotic TE[6], especially at the level of the DDE motif and the position of an upstream HTH. They also have comparable lengths and similar IR. The Merlin Tpase is longer at the N-terminus than that of IS1016 and more similar in size to the other members of the IS1595 family although it does not exhibit the IS1595 N-terminal ZF.
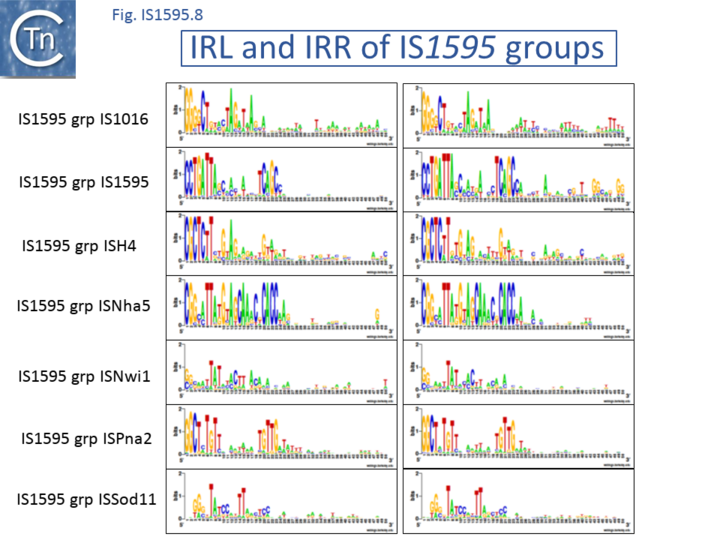
In the following, the different groups are described in an order according to their degree of divergence from IS1. Their ends are shown in Fig.1595.8.
Mechanism
There is no information at present concerning the mechanism of transposition of this group of elements.
ISPna2 group
The founder member of this group is from Polaromonas naphthalenivorans CJ2. It includes 18 members of slightly more than 1000 bp. Eleven resemble classical ISs, but the others carry downstream passenger genes or non-coding DNA (e.g. ISBse1) (Table IS1 1 and Table IS1 2). Like IS1 family members with a similar organization, these appear in single copies, raising the question of whether they are active. All members have IRs including a conserved terminal sequence similar to IS1 (Table IS1 1, Fig. IS1.4). ISSag10 (from Streptococcus agalatiae, now annotated as tISSag10 to indicate the presence of a passenger gene) is active. It includes a lincomycin resistance gene and, surprisingly, a functional origin of conjugative transfer embedded in the resistance gene. It is mobilizable by Tn916-type conjugative transposons[5]. A more complex partial derivative carrying potential multiple antibiotic resistance genes was identified in partially sequenced Campylobacter jejuni plasmid pCG8245[7] and includes typical IRs and an 8 bp AT-rich DR.
Several examples were also identified in the Bacilli. tISBwe1 from B. weihenstephanensis carries a 102 aa orf of unknown function, also present as an isolated orf (not associated with a transposon) in Bacillus cereus and Bacillus pumilus (Table IS1 1 and Table IS1 2). In a second IS copy, located on plasmid pE33L466 of B. cereus, much of the transposase has been deleted by recombination between two directly repeated TGAAATGA sequences. There are also partial copies in several other Bacillus species. ISBse1 from Bacillus selenitireducens includes 505 bp with no significant coding capacity. tISBsp1 (Bacillus sp.SG-1) also includes a 102 aa orf of unknown function which would be expressed convergently with the transposase. A second, isolated gene copy occurs in the SG-1 genome. A MITE (Miniature Inverted Repeat Transposable Element) derivative of 428 bp lacking coding sequences was also identified in Bacillus sp. NRRL B-14911. The Clostridia also carry members with passenger genes: a potential dihydrofolate reductase in tISCac2 (C. acetobutylicum ATCC824); two orfs of 235 aa and 115 aa with no known functions in tISCba1 (Clostridium bartelettii DSM16795); and an orf of unknown function of 103aa in tISClph1 (Clostridium phytofermentans) with an isolated homologue in Clostridium beijerinckii NCIMB8052. In contrast to tISSag10, in none of these cases has the presence of transfer functions been examined.
ISH4 archaeal group
This group contains only 4 members [4] with multiple and partial copies. They have a length of approximately 1000 bp. The IRs do not resemble those of IS1 or ISPna2 (Table IS1 1). At present, members of this group are restricted to the Halobacteriales and none include additional DNA.
IS1016 group
Another group of 19 different ISs which separates from the IS1595 is the IS1016 family. The IRs resemble those of IS1 and ISPna2 (Table IS1 1, Fig. IS1.4): These ISs are short (~700 bp). IS1016 itself, present in multiple copies (>20) in Neisseria gonorrhoeae and N. meningitidis and therefore likely to be active copies [3], does not carry an intact DDEK motif. Instead, all copies include a GDGK in place of the consensus (Fig.1595.1) (Table IS1 1). There is, however a potential E residue located further downstream as in several other groups. These genomes also carry many partial IS1016 copies. At present, members are limited to pathogenic bacteria of both plants and animals. For example, it is associated with an adhesin gene in invasive nontypeable Haemophilus influenzae[8].
IS1595 group
IS1595 group members are also about 1000 bp in length and include a single orf terminating within the right IR (Table IS1 1). The IRs do not resemble those of IS1 or ISPna2 (Fig. IS1.4). IS1595 has undergone expansion in X. oryzae to over 100 copies although there are only a few in the related X. campestris.
ISSod11 group (Fig.1595.1 and Fig.1595.2) (Table IS1 1 and Table IS1 2) members are about 1000 bp long with a single orf. The IRs show some variation in the terminal three nucleotides, but retain significant similarities possibly representing the transposase binding sequence in a subterminal region (Fig. IS1.4). No derivatives with passenger genes have been identified.
ISNwi1 group
Together with the ISNha5 group, these are perhaps the most complex. Of 16 identified members, all except two, ISPepr2 and ISRm32, include DNA sequences in addition to the transposase orf and all are present in only a single copy (Table IS1 2). Three of these are shown in Fig.1595.9 A. and a more detailed list can be found in Table IS1 2. There are three types: those carrying additional DNA of 600-1300 bp with no obvious orfs (e.g. ISNwi1); those carrying potential passenger genes downstream of the transposase orf (tISNwi2); and those carrying passenger genes before and after the transposase orf (tISBun1). These elements are between 1700 and 4000 bp (Table IS1 1 and Table IS1 2). Another complex sequence with a transposase highly similar to that of tISNwi2 (89% at the DNA level) and with similar IRs was identified in Nitrobacter hamburgensis (CP000319; 1803501-1814463). This has not been assigned a name since its transposase is degenerate; it is extremely long (10.9 kb) and carries at least 10 orfs including a large segment of bacteriophage Mu.
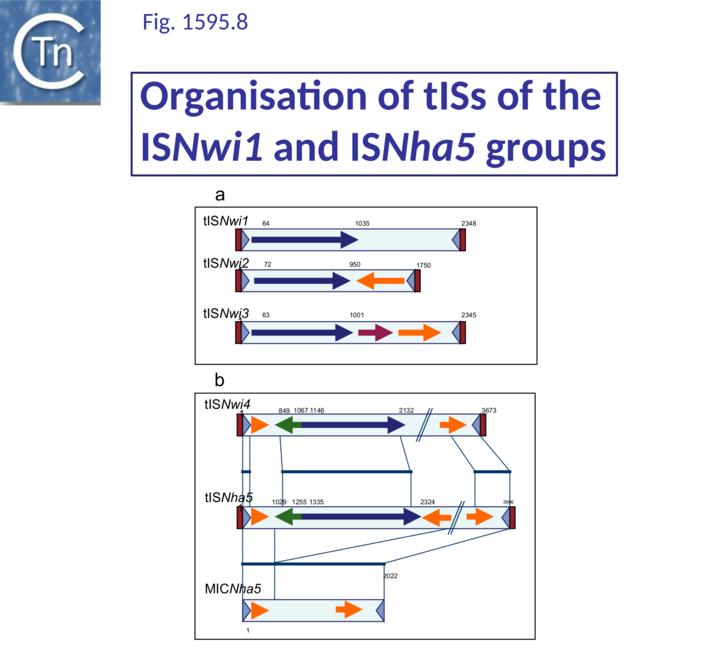
ISNha5 group
This group includes members which may carry passenger genes or additional non-coding DNA both upstream and downstream of the transposase orf (Table IS1 2; Fig.1595.9 B). Although from a structural and organizational point of view, these are quite diverse, their IRs are similar. All include a small divergently oriented orf located about 50 bp upstream from the transposase orf. In most cases, the product would be between 70 and 75 aa and resembles the HTH_XRE (or cro/C1-type HTH DNA-binding domain) proteins, a large family related to the cro and C1 repressors of temperate bacteriophages 434 and lambda whose members are DNA binding proteins involved in transcription control. This may represent a control element in transposase expression. A similar protein has been identified in IS231-derivative transposons which are unrelated to the ISNha5 group[9]. Two very similar members are found at the same position in the glycosyltransferase gene of Nitrobacter winogradskyi Nb-255 (tISNwi4) and N. hamburgensis X14 (tISNha5). These share significant DNA similarity but differ somewhat in their passenger genes (Fig.1595.9 B). A transposase-less MIC (Mobile Insertion Cassette [9]) derivative was also identified. A member from Azorhizobium caulinodans (tISAzca1) has an identical arrangement of HTH_XRE and transposase but the additional DNA upstream (983 bp) and downstream (2039 bp) appears to be non-coding (Table IS1 2). Other members carry another small orf with approximately the same size, orientation and distance from the transposase and which is also related to a transcriptional regulatory factor (Table IS1 2).
Bibliography
- ↑ Enright AJ, Van Dongen S, Ouzounis CA . An efficient algorithm for large-scale detection of protein families. - Nucleic Acids Res: 2002 Apr 1, 30(7);1575-84 [PubMed:11917018] [DOI]
- ↑ 2.0 2.1 Siguier P, Gagnevin L, Chandler M . The new IS1595 family, its relation to IS1 and the frontier between insertion sequences and transposons. - Res Microbiol: 2009 Apr, 160(3);232-41 [PubMed:19286454] [DOI]
- ↑ 3.0 3.1 Parkhill J, Achtman M, James KD, Bentley SD, Churcher C, Klee SR, Morelli G, Basham D, Brown D, Chillingworth T, Davies RM, Davis P, Devlin K, Feltwell T, Hamlin N, Holroyd S, Jagels K, Leather S, Moule S, Mungall K, Quail MA, Rajandream MA, Rutherford KM, Simmonds M, Skelton J, Whitehead S, Spratt BG, Barrell BG . Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. - Nature: 2000 Mar 30, 404(6777);502-6 [PubMed:10761919] [DOI]
- ↑ 4.0 4.1 Filée J, Siguier P, Chandler M . Insertion sequence diversity in archaea. - Microbiol Mol Biol Rev: 2007 Mar, 71(1);121-57 [PubMed:17347521] [DOI]
- ↑ 5.0 5.1 Achard A, Leclercq R . Characterization of a small mobilizable transposon, MTnSag1, in Streptococcus agalactiae. - J Bacteriol: 2007 Jun, 189(11);4328-31 [PubMed:17416666] [DOI]
- ↑ Feschotte C . Merlin, a new superfamily of DNA transposons identified in diverse animal genomes and related to bacterial IS1016 insertion sequences. - Mol Biol Evol: 2004 Sep, 21(9);1769-80 [PubMed:15190130] [DOI]
- ↑ Nirdnoy W, Mason CJ, Guerry P . Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. - Antimicrob Agents Chemother: 2005 Jun, 49(6);2454-9 [PubMed:15917546] [DOI]
- ↑ Satola SW, Napier B, Farley MM . Association of IS1016 with the hia adhesin gene and biotypes V and I in invasive nontypeable Haemophilus influenzae. - Infect Immun: 2008 Nov, 76(11);5221-7 [PubMed:18794287] [DOI]
- ↑ 9.0 9.1 De Palmenaer D, Vermeiren C, Mahillon J . IS231-MIC231 elements from Bacillus cereus sensu lato are modular. - Mol Microbiol: 2004 Jul, 53(2);457-67 [PubMed:15228527] [DOI]