IS Families/IS607 family
Contents
General
The first member of what is now called the IS607 family was IS1535 which had been identified as one of a collection of IS in the genome of Mycobacterium tuberculosis H37Rv [1]. Indeed, this study identified six different but related IS elements, IS1535 to IS1539 and IS1602, which, at the time, were grouped into a new IS family called the IS1535 family.
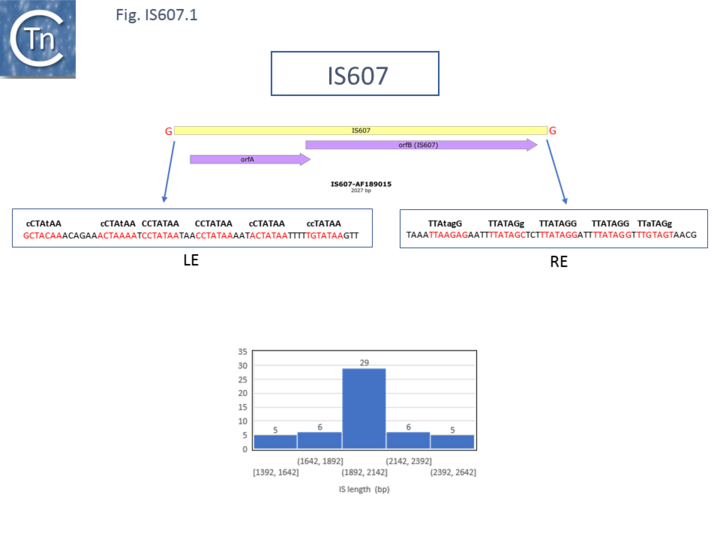
IS607 itself (Fig. IS607.1) was first identified by subtractive hybridization of a collection of Helicobacter pylori strains from a number of geographic locations and characterized by the Berg lab [2]. Further tests showed that it was widespread and present about 20% of H. pylori strains worldwide. It is 2027bp long and had a similar organization to IS1535. A second member of this family, IS609 (ISHp609), is also present in many H. pylori strains [3].
Distribution
Apart from being widespread geographically in Helicobacter strains [2] and in Mycobacteria [1][4]. Full length IS607 was widely distributed in many Helicobacter strains isolated from Africa, the Americas, Europe and India but only 1% East Asian strains [3]. IS607 family members have been subsequently found in a wide range of bacterial species, including cyanobacteria [5], and archea [6] and related sequences have been found in eukaryotic genomes and viruses [7][8], probably primarily through horizontal DNA transfer events.
One major problem in identifying IS607 family members is that, like members of the IS200/IS605 family, there are IS607 family members which have lost either orfA or, like IS200 itself, orfB. In the absence of clear sequence signatures which define the IS ends or of empty sites, it is difficult to define such reduced IS607 derivatives.
Organization
Most full-length IS607 family members are between 1900 and 2150 bp long (Fig. IS607.1) and carry two overlapping orfs where the stop codon from the upstream orf overlaps the start codon of the downstream orf suggesting that expression of the downstream gene is translationally coupled to that of the upstream gene [1] (see [9]) although IS607 was reported to encode to additional small orfs upstream of tnpA [2][3].
It was also noted that the product of the upstream orf, TnpA, shared similarity with Serine-site-specific Recombinases (SR) while that of the downstream frame, TnpB, showed weak similarity with TnpB of other IS such as IS1136 and IS891 [1][10][11] which do not include an upstream frame similar to orfA and which, due to the characteristic potential secondary structures at their ends, have been placed in the IS200/IS605 family.
The terminal DNA sequences of family members are not related although they carry several short directly repeated sequences at each end which appear to be helically phased [12][13] and a may contain a longer imperfect inverted repeat sequence near the ends but at different distances [12][13] (Fig. IS607.1 and Fig. IS607.3). Fig. IS607.2 shows the collection of ends described in reference [12] in green and reference [13] in black bold, It was noted that many, but not all, IS607 family members in ISfinder carry a trinucleotide repeated at each end (shown in reference [12], Fig. IS607.2 ) which may be involved in the transposition reaction [12]. The absence of these in some examples may simply be due to an incorrect definition of the IS ends.

Mechanism
IS607 was observed to transpose in Escherichia coli, and the sequence of the IS-target junctions showed that IS607 inserted into a GG dinucleotide (Fig. IS607.1; see also Fig. IS607.2). Insertion sometimes generated a 2 bp target repeat but in other cases, no target repeat was observed. Mutational studies demonstrated that TnpA was required for transposition [2] but, as in the case in members of the IS200/IS605 family TnpB is not. IS609 was found to insert in many chromosomal sites and, in contrast to IS607, to have a preference for insertion with its left end next to a trinucleotide, TAT [3].
As pointed out by Boocock and Rice [12], the IS607 transposase is unusual for a serine site-specific recombinase because most such enzymes require extensive sequence-specificity for all recombining partners [14][15]. As might be expected for a transposase whose function is to optimise the number of potential target sites, IS607 TnpA does not [3][16]. Integration does not require a large conserved target sequence.
Serine recombinases are generally of two types: small (smSR) which orchestrate highly regulated recombination reactions such as the Tn3 transposon family, resolution or inversion of invertible DNA segments such as Hin and Gin [17][18][19], and large SR often involved with phage integration [20]; Both carry their DNA binding domains at the C-terminus rather than TnpA of IS607 family members with their DNA binding domain at the N-terminus.
The reaction pathway of well characterized smSR has been studied in exquisite detail. It involves a recombinase tetramer and both DNA recombination partners with multiple recombinase binding sites forming a precise architectural structure. However, the N-terminal DNA binding domain in IS607 family transposases would be expected to be incompatible with tetramer formation on the present structural models [12][13][18][19][21].
DNA at the recombining sites is cleaved by attack of the conserved catalytic serine nucleophile to generate a 3′ hydroxyl and a covalent 5’ phospho-serine protein-DNA intermediate. Following cleavage, the two recombinase subunits of the tetramer attached to the 5’ DNA ends (the cutting dimer[12]) are rotated 180° by the other two (the “rotating” dimer [12]) [22][23][24][25][26][27][28]. The “opposing” 3 ′ hydroxyl groups then attack the phosphoserine linkages of the rotated partners to reform a phosphodiester bond and complete the strand transfer reaction. The GG dinucleotide at the transposon termini (Fig. IS607.2, blue underlined) and the invariant GG at the insertion target sites of these IS, might represent the 2 bp “overlap” sequence observed at the recombination crossover site for other serine recombinases [12][13].
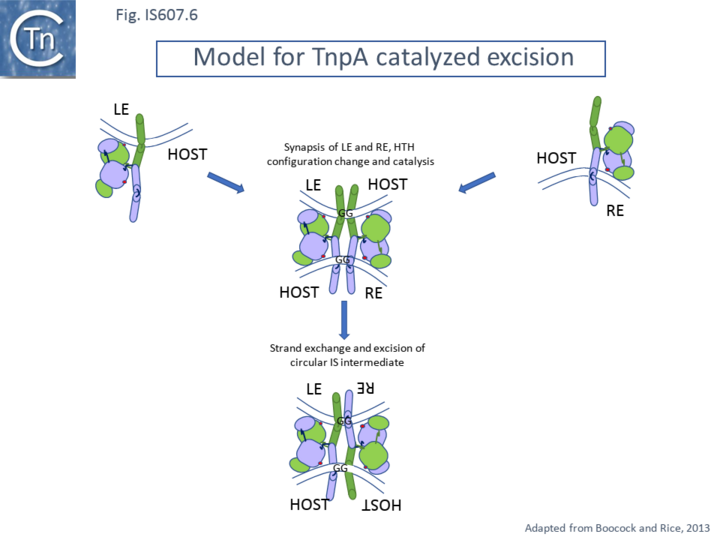
Like many IS, it is thought that IS607 transposases using a double strand closed circular intermediate as judged by the presence of a LE-RE junction identified by PCR in an E. coli transposition system (Grindley, personal communication and cited in ref [12]) in which the IS is excised from its donor site with both ends joined and the donor DNA is resealed [12]. This model is reasonable in view of the type of transposase involved but remains to be formally validated.
The Johnson laboratory [13] has investigated the transposases of three related IS607 family members: IS607 itself, IS1535, and ISC1926 from Sulfolobus islandicus, a hyperthermophilic archea [29]. Using a lambda hop assay [30] they confirmed that IS607 indeed inserts between the G residues in a GG dinucleotide target; that transposition requires an intact TnpA, mutation of the active site serine (vertical arrow in Fig. IS607.2) with glycine eliminated activity. While TnpA promoted transposition in this assay, a construct supplying TnpA and TnpB did not and it was concluded that TnpB inhibits transposition. This is similar to the observed effect of the related TnpB on transposition of the IS200/IS605 family member, ISDra2 [31].
Purified TnpA from all three IS, IS607 itself, IS1535, and ISC1926 was able to bind to the cognate IS ends in gel mobility shift assays (EMSA) [13]. More extensive studies, directed to TnpA of IS1535 (TnpAIS1535) due to its more robust binding activity, using EMSA, DNase footprinting and exonuclease digestion, indicated that: it binds cooperatively to multiple sites (the directly repeated sequences) in LE and can bind a second LE to form a Paired End Complex (PEC); TnpA “nucleation” occurs over four of the helically-phased 9 bp direct repeat LE motifs (Fig. IS607.2); LE motifs (a) through part of (d) are required for PEC formation, but efficiency is improved by including “non-specific” DNA both within LE and at the IS-host DNA junction; RE is a poor substrate for TnpA binding, possibly resulting from the lower number of repeat motifs; TnpA covered only the two repeated RE motifs and supported only a low level of RE-RE or RE-LE PEC formation; although protected from exonuclease digestion, no clear footprint could be detected on RE indicating a lower binding affinity [13]. Unpublished data from these authors citing binding studies with TnpAIS607 and TnpAIS1926 however, suggested that this is not a general property of all members of the IS607 family since neither exhibited such large differences in the capacity to bind LE and RE ends of their cognate elements.
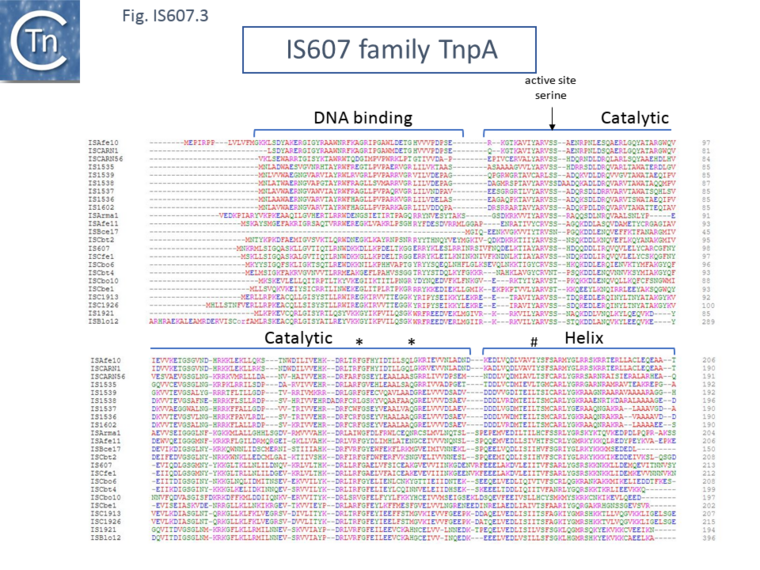
Further functional analysis of TnpAIS1535 revealed that removal of the 50 N-terminal amino acids eliminated binding activity [13], confirming its role in TnpA function [12]. When well conserved residues in the catalytic domain (* in Fig. IS607.3) were substituted for cysteine, the capacity of the mutant proteins to generate PECs under oxidizing (cross linked) or reducing conditions was identical. This was interpreted as showing that there are no large-scale conformational changes in the catalytic domain on binding. However, a similar substitution in the C-terminal HTH domain (# in Fig. IS607.3) eliminated PEC formation when oxidized as did deletion of this region, suggesting that binding involves conformational changes in this domain.
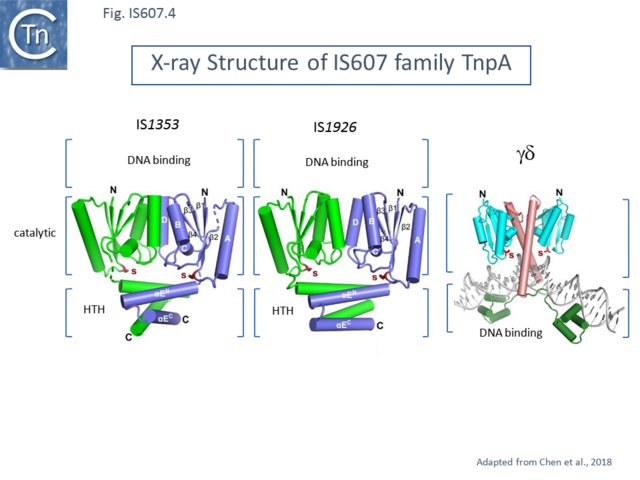
Structural analysis also underlined the difference between IS607 family TnpA and other serine recombinases. Structures were solved for the combined catalytic and C-terminal HTH domains of TnpAIS1535 and of TnpAIS1926 [13] (Fig. IS607.3) and for TnpAISC1904 from Sulfolobus solfataricus P2 with similar results [12]. The structures were found to contain either one (TnpAIS1535) or two TnpAIS1926 dimers. The topology of the catalytic domains was identical to that of the catalytic core of the smSR catalytic domain.
However, whereas the dimer interface of smSR occurs between the long helices at the C-terminal end of the catalytic domain, that of both TnpAIS1535 and of TnpAIS1926 is located between entirely different helices in the catalytic domain (Fig. IS607.4). It was proposed that this C-terminal HTH domain must be displaced to permit DNA to enter the active site. Moreover, since the DNA binding domain is located on the opposite side of the dimer to the active site, it seemed probable that cleavage occurs in trans (where a molecule bound to one recombining pair cleaves the other partner) [13] as suggested in the model proposed by Boocock and Rice [12], a characteristic of many transposable elements (see [32][33][34] for reviews).
The unusual dimer structure originally noted for TnpAISC1904 led to an elegant detailed mechanistic model[12]. In the Boocock-Rice model, which addressed both integration and excision of the proposed circular IS intermediate, a TnpA tetramer forms a complex with both recombining DNA partners. For integration the tetramer assembles on the IS circle LE-RE junction (Fig. IS607.4) while in excision, it is proposed to assemble from dimers bound a each end during synapsis (Fig. IS607.5 and Fig. IS607.6) at it is proposed that binding of the dimer occurs to LE (or RE) in the circle junction using TnpA dimer (B) using its DNA binding domain. For catalysis to occur, the active site must be “demasked” (Fig. IS607.4 top). The TnpAISC1904 structure indicated that the C-terminal HTH domain must be able to move to render the catalytic center accessible to DNA[12] and this was confirmed by crosslinking experiments indicating that the C-terminal HTH domain must be able to move for PEC formation however, cross linking of the catalytic domain did not affect PEC formation. A second dimer is then proposed to bind to the RE (or LE) side of the junction leading to binding of the insertion site and its engagement in the catalytic site. The model proposes that the free DNA binding DNA of a single dimer is not capable of binding DNA non-specifically on its own but two such domains in the tetramer are proficient in non-specific DNA binding and the tetramer bound to the IS junction can thus accommodate the relatively non-specific target DNA.
To understand the mechanism of IS607 family transposition in detail (for example at which stage in the pathway the HTH configuration change occurs, the stoichiometry of the transpososome, how the different repeated sequence elements bind TnpA) will require further structural studies using DNA-protein cocrystals.

TnpAS IS607 Excision Activity
An in vivo reaction has been developed to monitor IS607 excision [35]. This was undertaken in a study of IStrons, group I self-splicing introns in which include members of the IS220/IS605 or IS607 families whose activities are intimately associated with the splicing and RNA guide endonuclease (TnpB) activities of the “host” IStron.
Žedaveinytė et al 2023 [35] used a two-plasmid experimental system in E.coli with a mini-IS607 transposon derived from an IStron, CboIStron from C. botulinum strain BKT015925 located on a large botulinum neurotoxin-encoding plasmid. CboIStron carries full length tnpAS and tnpB genes (Fig. IS200.59). The mini-IS607 included 60bp from each IS end but lacked both TnpAs and TnpB genes (Fig.IS607.7). Excision products were assayed by qPCR following transformation of a strain carrying a TnpAs-producing plasmid with the mini-IS607 donor plasmid and growth overnight (Fig.IS607.7). They occurred at a frequency of 5% and required a functional TnpAS active site and both IStron(IS) ends and carried a precise donor joint. The authors indicate that this several orders of magnitude higher than that catalyzed by TnpAY in an IS605 system under similar conditions; [36]. Internal end deletions showed a requirement for 40 (LE) and 60 (RE) bp for robust excision. In particular, the ends carry a subset of the repeated elements (3 in LE and 2 in RE) of those identified by Boocock and Rice [37] and Chen et al. [38] (Fig. IS607.2) and implicated in the cooperative assembly of multiple TnpAS dimers into a synaptic complex [38]. Mutation of these repeats individually resulted in reduced excision activity while multiple mutations eliminated excision entirely [35]. Circular copies of the mini-IS607 were also detected which had abutted LE and RE ends.
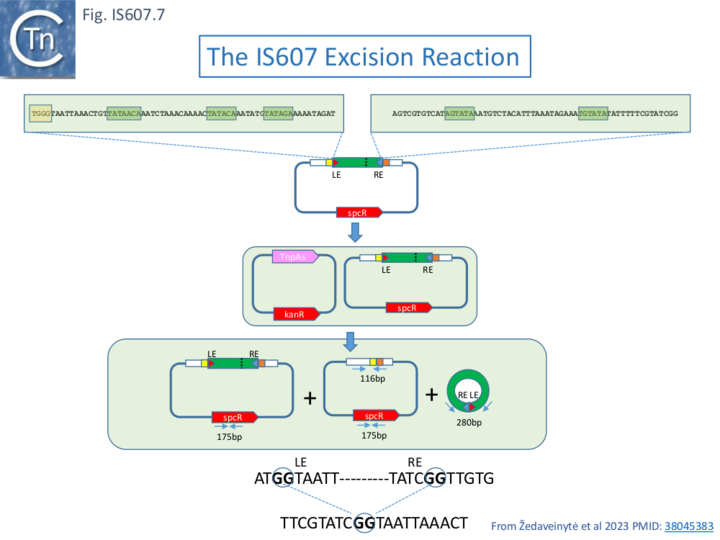
IS607 Integration
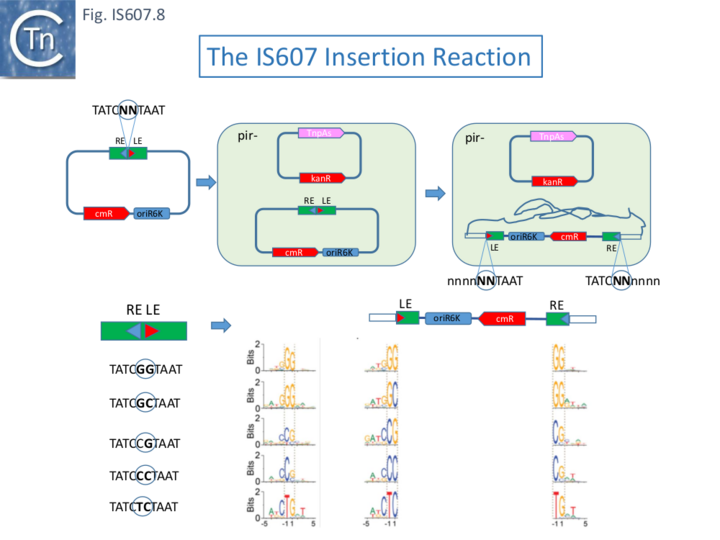
TnpAS IS607 Insertion Activity
To investigate IS607 insertion, a chloramphenicol resistance tagged suicide (non-replicative) donor plasmid carrying abutted LE and RE and a pir-dependent R6K replication origin, ori, was used: grown in a pir+ strain and transformed into a pir- strain expressing TnpAS. Cell viability in the presence of chloramphenicol was dependent on TnpAS (Fig.IS607.8). Integration specificity was investigated by genome-wide sequencing. A GG or GC nucleotide was present between the abutted LE and RE. Integration was found to require require a GG dinucleotide target with a preference, but not an absolute requirement, for the sequence: TGGG (this is the predicted TAM sequence - see below). Though rare, non-canonical core nucleotides, GC, could be used in integration. In these cases, the GC dinucleotide was present at the LE end while GG occurred at RE. However, other rare examples are more difficult to explain (Fig.IS607.8).
TnpB and maintenance of the TE
IS607 family members also carry a TnpB gene similar to that found in IS605 family members. Much of what is known of these comes from studies of IStrons [35] and of ISXfa1, an IS derivative from Xylella fastidiosa[39] .
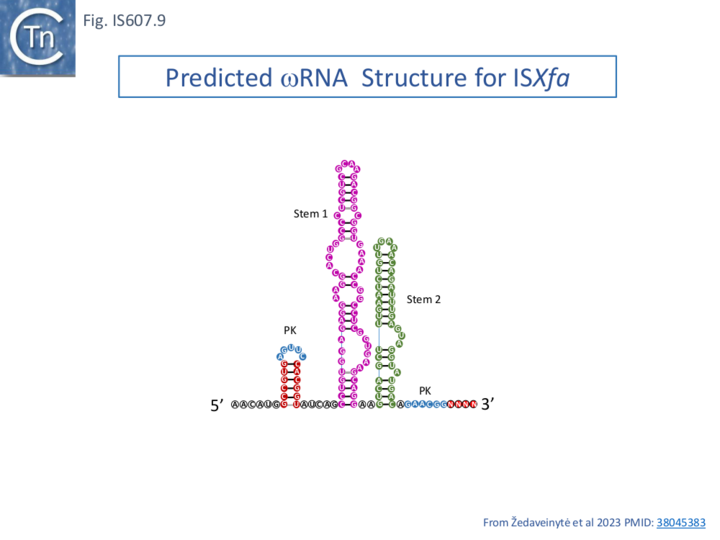
Analysis of small-RNA expressed from ISXfa1 in E. coli identified ∼ 150 nt reRNA (an RNA produced from the right IS end and extending beyond the transposon boundary) consistent with a guide RNA (re or ωRNA) used by TnpB guide endonucleases to cleave a target DNA next to the transposon associated motif at the left end (the TAM sequence) and, using co-variance models [35] to assume a specific secondary structure typical of guide-RNAs (Fig. IS6007.9 see also Fig. IS200.62). In view of their close evolutionary and functional similarities, a more extensive treatment of TnpB/re(ω) RNA activities of both IS605 and IS607 is provided in the IS200/IS605 chapter section TnpB.
Preventing IS607 Extinction.
Like members of the IS200/IS605 family (Fig. IS200.55) IS607 family transposition results in loss of the transposon from the donor site (Fig. IS607.7 and Fig. IS607.8) and could therefore lead to IS extinction. The major difference between the two IS families is that for IS605, excision of a single strand circle occurs from the lagging strand to form a single-strand circle but leaving an intact copy in the leading strand. The RNA guide together with TnpB serves to replace the excised copy, presumably at the replication fork (Fig. IS200.55) linking transposition to host replication and preventing IS extinction [36]. Integration also appears to occur at the replication fork into the lagging strand resulting in insertion of the IS in a specific orientation.
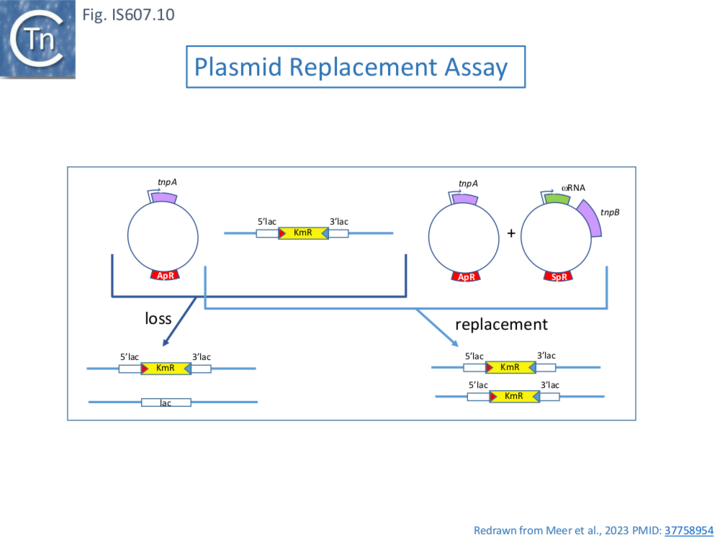
In the case of IS607, it was shown that TnpB/re(RNA prevents IS loss but does not affect transposition activity. This was tested using the retention/replacement assay of Meers et al [36](Fig IS607.10 see also Fig. IS200 54B) where an IS derivative interrupted a plasmid-based lacZ gene and oriented opposite to the direction of transcription of the gene (to avoid the splicing reaction as has been used in assays of transposition of retroelements. Expression of TnpAS alone resulted in transposon loss evidenced by about half lac+ colonies, a frequency reduced about 10x by co-expression of wildtype but not mutant TnpB. The veracity of these results was confirmed by PCR [35].
While IS replacement for members of the IS605 group is conceptually easy to understand in the context of a coupling to the host replication fork, it is more difficult to understand how replacement of IS607 family members might occur unless their transposition is also linked in some way to replication. Such linkage might not be physical but, like that of Tn10/IS10 and Tn5/IS50 transposition [41][42], controlled by passage of a replication fork: in these cases, by the state of methylation of DAM sites in the IS ends (Fig. IS4.4, Fig. IS4.5). Other signals resulting from passage of a replication fork could be imagined to be used to couple transposition. Additionally, the IS might be copied to fill the empty site from a daughter “chromatid” in multifork replicating chromosomes.
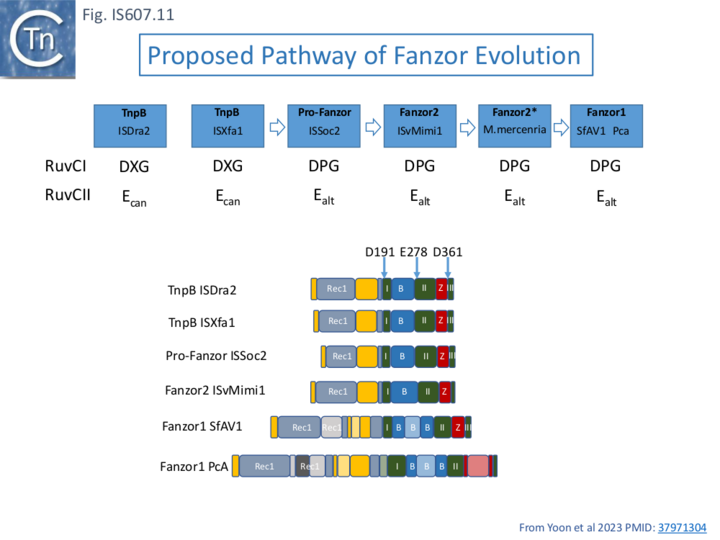
Is IS607 TnpB the Ancestor of Fanzor Proteins?
Fanzor proteins, Fanzor1 and Fanzor2, are somewhat longer eukaryotic relatives of TnpB [43]. These have also been shown to be associated with guide RNA and to function as RNA-guided endonucleases.
Extensive database mining phylogenetic analysis and structural modeling [44] have led to the suggestion that TnpB from an IS607 ancestor was a precursor to Fanzor2 which in turn gave rise to Fanzor1 (Fanzor2 and/or Fanzor1 may have evolved from an IS607 ancestor (Fig IS607.11). For a more detailed explanation see "The Functional Relationship Between Fanzor Evolution and IS607 TnpB" in the IS200/IS605 section.
Bibliography
- ↑ 1.0 1.1 1.2 1.3 <pubmed>10220167</pubmed>
- ↑ 2.0 2.1 2.2 2.3 <pubmed>PMC110970</pubmed>
- ↑ 3.0 3.1 3.2 3.3 3.4 <pubmed>PMC524915</pubmed>
- ↑ <pubmed>9634230</pubmed>
- ↑ <pubmed>21576885</pubmed>
- ↑ <pubmed>PMC1847376</pubmed>
- ↑ <pubmed>PMC3673617</pubmed>
- ↑ <pubmed>17109990</pubmed>
- ↑ <pubmed>2183416</pubmed>
- ↑ <pubmed>8386127</pubmed>
- ↑ <pubmed>PMC210459</pubmed>
- ↑ 12.00 12.01 12.02 12.03 12.04 12.05 12.06 12.07 12.08 12.09 12.10 12.11 12.12 12.13 12.14 12.15 12.16 <pubmed>PMC4058570</pubmed>
- ↑ 13.00 13.01 13.02 13.03 13.04 13.05 13.06 13.07 13.08 13.09 13.10 <pubmed>PMC6188088</pubmed>
- ↑ <pubmed>16756503</pubmed>
- ↑ <pubmed>20298189</pubmed>
- ↑ <pubmed>PMC134827</pubmed>
- ↑ Grindley NDF. The Movement of Tn3-Like Elements: Transposition and Cointegrate Resolution. In: Craig NL, Lambowitz AM, Craigie R, Gellert M, editors. Mobile DNA II. American Society of Microbiology; 2002. p. 272–302.
- ↑ 18.0 18.1 <pubmed>PMC4384473</pubmed>
- ↑ 19.0 19.1 <pubmed>PMC5659196</pubmed>
- ↑ <pubmed>26350324</pubmed>
- ↑ <pubmed>26104451</pubmed>
- ↑ <pubmed>PMC397197</pubmed>
- ↑ <pubmed>1885004</pubmed>
- ↑ <pubmed>8757793</pubmed>
- ↑ <pubmed>6313351</pubmed>
- ↑ <pubmed>2548736</pubmed>
- ↑ <pubmed>2990045</pubmed>
- ↑ <pubmed>15994378</pubmed>
- ↑ <pubmed>15612937</pubmed>
- ↑ <pubmed>PMC1213899</pubmed>
- ↑ <pubmed>23461641</pubmed>
- ↑ <pubmed>PMC7292550</pubmed>
- ↑ <pubmed>PMC3107681</pubmed>
- ↑ <pubmed>26104718</pubmed>
- ↑ 35.0 35.1 35.2 35.3 35.4 35.5 35.6 35.7 35.8 <pubmed>38045383</pubmed>
- ↑ 36.0 36.1 36.2 36.3 <pubmed>37758954</pubmed>
- ↑ <pubmed>24195768</pubmed>
- ↑ 38.0 38.1 <pubmed>30289389</pubmed>
- ↑ <pubmed>37971304</pubmed>
- ↑ <pubmed>37386294</pubmed>
- ↑ <pubmed>3000598</pubmed>
- ↑ <pubmed>2451025</pubmed>
- ↑ <pubmed>23548000</pubmed>
- ↑ <pubmed>37971304</pubmed>