Difference between revisions of "Transposons families/compound transposons"
| Line 54: | Line 54: | ||
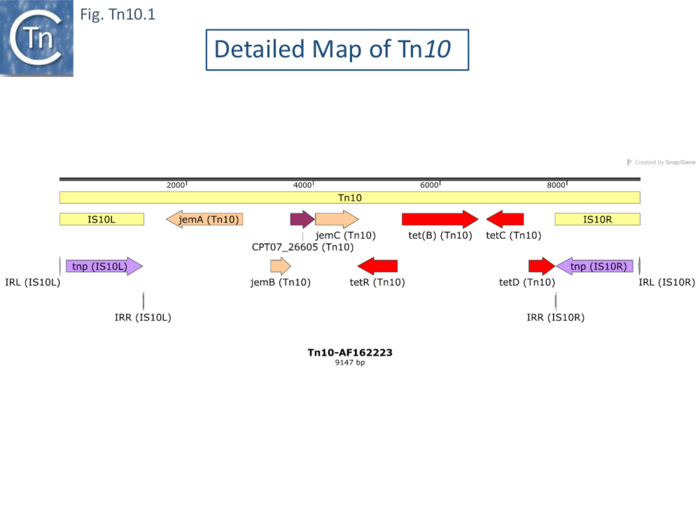
Five of 26 deletions (#250, 260, 261 264 and 317; [[:File:FigTn10-3.png|Fig. Tn10.3 '''A''']]) appeared to have retained the flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] and were missing parts of the interior [[wikipedia:Tetracycline|''tet'' region]]. All had deletion endpoints which mapped to one or other or, in the case of #317, both of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] '''IE'''. Three were found to have deleted one or other of the flanking IS either partially (#267) or fully (#270, 265). | Five of 26 deletions (#250, 260, 261 264 and 317; [[:File:FigTn10-3.png|Fig. Tn10.3 '''A''']]) appeared to have retained the flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] and were missing parts of the interior [[wikipedia:Tetracycline|''tet'' region]]. All had deletion endpoints which mapped to one or other or, in the case of #317, both of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] '''IE'''. Three were found to have deleted one or other of the flanking IS either partially (#267) or fully (#270, 265). | ||
| − | The second class of deletion was accompanied by small inversions. Six deletion derivatives were analysed in detail ( [[:File:FigTn10-3.png|Fig. Tn10.3 '''B''']]). In all, the intervening DNA between the flanking IS was deleted. These types of deletion and inversion/deletion have been proposed <ref name=":16" /> to occur by intramolecular transposition using both flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10'']'''IE''' (Fig. Comp. | + | The second class of deletion was accompanied by small inversions. Six deletion derivatives were analysed in detail ( [[:File:FigTn10-3.png|Fig. Tn10.3 '''B''']]). In all, the intervening DNA between the flanking IS was deleted. These types of deletion and inversion/deletion have been proposed <ref name=":16" /> to occur by intramolecular transposition using both flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10'']'''IE''' ([[:File:Fig-Comp1.png|Fig. Comp.1]]) deletions occur when insertion occurs into the target in one orientation ([[:File:Fig-Comp1.png|Fig. Comp.5 '''left''']]) while inversion/deletions occur when insertion occurs in the opposite orientation ([[:File:Fig-Comp1.png|Fig. Comp.5 '''left''']]). The inversion/deletion events should give rise to circular products of the deleted DNA but, since they are unable to replicate, are lost during cell growth. Such circular products had subsequently been observed <ref><nowiki><pubmed>6091910</pubmed></nowiki></ref>. They proved to be interrupted in one or other of the DNA strands and were interpreted as abortive transposition events in which the transposon had inserted within itself producing abutted left and right ends. A later study <ref><nowiki><pubmed>2553269</pubmed></nowiki></ref> demonstrated that these were in fact deletion/inversion products in which the transposon ends had inserted in targets within the transposon. |
The third type of ''[[wikipedia:Tetracycline|tet<sup>S</sup>]]'' deletion, analyzed in an associated study was called “nearly precise excision” ( [[:File:FigTn10-3.png|Fig. Tn10.3 '''C''']] ) <ref><nowiki><pubmed>455446</pubmed></nowiki></ref>. These appeared to have occurred at AT-rich sub-terminal inverted repeats located at the outside end ('''OE'''; [[:File:FigTn10-3.png|Fig. Tn10.3 '''C''']]) <ref name=":15" /> which are involved in the RNAin/RNAout antisense RNA regulation of transposase expression <ref><nowiki><pubmed>6311438</pubmed></nowiki></ref><ref><nowiki><pubmed>1691097</pubmed></nowiki></ref><ref><nowiki><pubmed>25649688</pubmed></nowiki></ref>. | The third type of ''[[wikipedia:Tetracycline|tet<sup>S</sup>]]'' deletion, analyzed in an associated study was called “nearly precise excision” ( [[:File:FigTn10-3.png|Fig. Tn10.3 '''C''']] ) <ref><nowiki><pubmed>455446</pubmed></nowiki></ref>. These appeared to have occurred at AT-rich sub-terminal inverted repeats located at the outside end ('''OE'''; [[:File:FigTn10-3.png|Fig. Tn10.3 '''C''']]) <ref name=":15" /> which are involved in the RNAin/RNAout antisense RNA regulation of transposase expression <ref><nowiki><pubmed>6311438</pubmed></nowiki></ref><ref><nowiki><pubmed>1691097</pubmed></nowiki></ref><ref><nowiki><pubmed>25649688</pubmed></nowiki></ref>. | ||
| Line 69: | Line 69: | ||
====“Precise” Excision.==== | ====“Precise” Excision.==== | ||
| − | Foster et al <ref name=":18"><nowiki><pubmed>6260376</pubmed></nowiki></ref> identified a number of [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10'']-mediated events in addition to the adjacent deletions and nearly-precise excision (Fig. Tn10.3). These, which all generate new joints between the transposon end and a target sequence, are precise excision of the entire [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] and precise excision of the [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] remnant remaining after nearly precise excision. The novel deletions and nearly-precise excision all involved small direct repeats at or near the [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] ends resulting in deletion of all intervening DNA but leaving one of the repeats in place. The DNA sequence of a ''his''+ revertant of a ''his''::[https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] derivative of ''[[wikipedia:Salmonella_enterica_subsp._enterica|S.typhimurium]]'' revealed a wildtype sequence with a single copy of the direct target repeat generated by the original [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] insertion event. The ''his'' system also generated nearly precise excision events using subterminal repeats in OE similar to those found in the lambda system (Fig.Tn10. | + | Foster et al <ref name=":18"><nowiki><pubmed>6260376</pubmed></nowiki></ref> identified a number of [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10'']-mediated events in addition to the adjacent deletions and nearly-precise excision ([[:File:FigTn10-3.png|Fig. Tn10.3]]). These, which all generate new joints between the transposon end and a target sequence, are precise excision of the entire [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] and precise excision of the [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] remnant remaining after nearly precise excision. The novel deletions and nearly-precise excision all involved small direct repeats at or near the [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] ends resulting in deletion of all intervening DNA but leaving one of the repeats in place. The DNA sequence of a ''his''+ revertant of a ''his''::[https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] derivative of ''[[wikipedia:Salmonella_enterica_subsp._enterica|S.typhimurium]]'' revealed a wildtype sequence with a single copy of the direct target repeat generated by the original [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] insertion event. The ''his'' system also generated nearly precise excision events using subterminal repeats in OE similar to those found in the lambda system ([[:File:FigTn10-3.png|Fig.Tn10.3 '''C''']]). These physical structures are consistent with previous genetic results in which a [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] insertion into ''hisG'' (''hisG''::[https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10'']) is polar on the downstream ''hisD'' <ref name=":17" />. Two types of ''hisD'' polar-relief revertants were obtained. In one class, the authors could find little or no [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] homologous material (with a resolution of >50 bp by electron microscopy) but these could subsequently give rise to full ''his''+ offspring. This can now be interpreted as a twostep process with an initial nearly precise excision, which leaves 50 bp of [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] ([[:File:FigTn10-3.png|Fig.Tn10.3 '''C''']]) and is non-polar on ''hisD'', followed by a precise excision of the remaining [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] material to generate a his+ phenotype.[https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223] |
| − | Moreover: [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] derivatives defective for transposition are unaffected in their excision and nearly precise excision frequencies; excision and nearly precise excision frequencies decrease with [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] deletion derivatives carrying shorter inverted repeats ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10'']) <ref name=":18" />. Similar conclusions were reached for [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''], which is structurally similar to [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] (Fig. Comp.1), strongly suggesting that long inverted flanking IS are key (for [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] see also Egner and Berg <ref><nowiki><pubmed>6264444</pubmed></nowiki></ref>). | + | Moreover: [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] derivatives defective for transposition are unaffected in their excision and nearly precise excision frequencies; excision and nearly precise excision frequencies decrease with [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] deletion derivatives carrying shorter inverted repeats ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10'']) <ref name=":18" />. Similar conclusions were reached for [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''], which is structurally similar to [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] ([[:File:Fig-Comp1.png|Fig. Comp.1]]), strongly suggesting that long inverted flanking IS are key (for [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] see also Egner and Berg <ref><nowiki><pubmed>6264444</pubmed></nowiki></ref>). |
Rearrangements promoted by a single chromosomal insertion of [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] or an artificial Tn''Gal'' in ''[[wikipedia:Escherichia_coli|E. coli]]'' and ''S. typhimurium'' were analyzed by [[wikipedia:Southern_blot|Southern blot hybridization]] of DNA isolated from many individual clones <ref><nowiki><pubmed>3038673</pubmed></nowiki></ref> and digested with the appropriate [[wikipedia:Restriction_enzyme|restriction enzymes]]. This provided physical evidence of transposition events by following the retention or loss of Tn-chromosome junctions and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10'']-internal transposon junctions. The authors were able to estimate transposition frequencies of the entire transposon and of the individual flanking IS: transposition of an individual [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] was 10<sup>-4</sup> per cell per generation in overnight cultures grown and plated on minimal media, a value at least ten fold higher than other [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10'']/[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] promoted events (e.g. deletions etc); all chromosomal rearrangements were either deletion/inversions or deletions as had been previously analyzed for [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10'']; the rearrangements were at least 10 fold lower than [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] transposition; and [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] events used nearby target sites while [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] events appeared to target “randomly” around the chromosome. | Rearrangements promoted by a single chromosomal insertion of [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] or an artificial Tn''Gal'' in ''[[wikipedia:Escherichia_coli|E. coli]]'' and ''S. typhimurium'' were analyzed by [[wikipedia:Southern_blot|Southern blot hybridization]] of DNA isolated from many individual clones <ref><nowiki><pubmed>3038673</pubmed></nowiki></ref> and digested with the appropriate [[wikipedia:Restriction_enzyme|restriction enzymes]]. This provided physical evidence of transposition events by following the retention or loss of Tn-chromosome junctions and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10'']-internal transposon junctions. The authors were able to estimate transposition frequencies of the entire transposon and of the individual flanking IS: transposition of an individual [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] was 10<sup>-4</sup> per cell per generation in overnight cultures grown and plated on minimal media, a value at least ten fold higher than other [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10'']/[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] promoted events (e.g. deletions etc); all chromosomal rearrangements were either deletion/inversions or deletions as had been previously analyzed for [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10'']; the rearrangements were at least 10 fold lower than [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] transposition; and [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] events used nearby target sites while [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] events appeared to target “randomly” around the chromosome. | ||
====Transposition or Replication?==== | ====Transposition or Replication?==== | ||
| − | It was suggested that these deletions may be generated during the transposition process when two [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10'']OE ends ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10L IS''10''L] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''R]) are synapsed which would explain why excision is less frequent than transposition. An alternative explanation would be that excision occurs during replication when a [[wikipedia:DNA_replication#Replication_fork|replication fork]] passes through the transposon. This might allow pairing of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10L IS''10''L] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''R] to form “snapbacks” (see Fig. 2.4) on the discontinuous (lagging) strand, facilitating replication slippage between the two directly-repeated sequences. This explanation has been proposed in the case of precise excision of [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5'']. | + | It was suggested that these deletions may be generated during the transposition process when two [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10'']OE ends ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10L IS''10''L] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''R]) are synapsed which would explain why excision is less frequent than transposition. An alternative explanation would be that excision occurs during replication when a [[wikipedia:DNA_replication#Replication_fork|replication fork]] passes through the transposon. This might allow pairing of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10L IS''10''L] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''R] to form “snapbacks” (see [[:File:1.2.4.png|Fig. 2.4]]) on the discontinuous (lagging) strand, facilitating replication slippage between the two directly-repeated sequences. This explanation has been proposed in the case of precise excision of [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5'']. |
<br /> | <br /> | ||
| Line 102: | Line 102: | ||
====Other Properties of Tn''10'': Active IE ends, Inverse Transposition==== | ====Other Properties of Tn''10'': Active IE ends, Inverse Transposition==== | ||
| − | That the inside ends are active in transposition as implied in the deletion data (Fig. Tn10.3) was also demonstrated by the observation that integrative suppression (the rescue of chromosome replication in a temperature sensitive ''dnaAts'' initiation mutant by plasmid [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 integration]) using the R100.1 plasmid (a mutant derivative of R100 derepressed for conjugal transfer) to integrate and drive chromosome replication <ref><nowiki><pubmed>328481</pubmed></nowiki></ref> resulted in the frequent appearance of [[wikipedia:Tetracycline|''tet'' sensitive]] temperature resistant clones (Chandler et al., 1978 CSH Symp. Quant. Biol. 43). Further analysis demonstrated that in these cells, R100.1 had integrated into the host chromosome by transposition using the '''IE''' of both [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] with the loss of ''[[wikipedia:Tetracycline|tetR]]''. This was termed “inverse” transposition <ref><nowiki><pubmed>381840</pubmed></nowiki></ref>. Similar use of '''IE''' was subsequently reported in the case of a circular plasmid carrying and ampicillin resistance gene together with a [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] which gave rise to two types of symmetrical transposition event: normal [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] transposition of [[wikipedia:Tetracycline|tet<sup>R</sup>]] using both '''OE''' and another involving [[wikipedia:Ampicillin|amp<sup>R</sup>]] and both opposite, '''IE''', ends in which ''[[wikipedia:Tetracycline|tet]]'' was lost <ref><nowiki><pubmed>6260375</pubmed></nowiki></ref>. | + | That the inside ends are active in transposition as implied in the deletion data ([[:File:FigTn10-3.png|Fig. Tn10.3]]) was also demonstrated by the observation that integrative suppression (the rescue of chromosome replication in a temperature sensitive ''dnaAts'' initiation mutant by plasmid [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 integration]) using the R100.1 plasmid (a mutant derivative of R100 derepressed for conjugal transfer) to integrate and drive chromosome replication <ref><nowiki><pubmed>328481</pubmed></nowiki></ref> resulted in the frequent appearance of [[wikipedia:Tetracycline|''tet'' sensitive]] temperature resistant clones (Chandler et al., 1978 CSH Symp. Quant. Biol. 43). Further analysis demonstrated that in these cells, R100.1 had integrated into the host chromosome by transposition using the '''IE''' of both [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] with the loss of ''[[wikipedia:Tetracycline|tetR]]''. This was termed “inverse” transposition <ref><nowiki><pubmed>381840</pubmed></nowiki></ref>. Similar use of '''IE''' was subsequently reported in the case of a circular plasmid carrying and ampicillin resistance gene together with a [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] which gave rise to two types of symmetrical transposition event: normal [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] transposition of [[wikipedia:Tetracycline|tet<sup>R</sup>]] using both '''OE''' and another involving [[wikipedia:Ampicillin|amp<sup>R</sup>]] and both opposite, '''IE''', ends in which ''[[wikipedia:Tetracycline|tet]]'' was lost <ref><nowiki><pubmed>6260375</pubmed></nowiki></ref>. |
====Other Properties of Tn''10'': The Flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10L IS''10''] are Functionally Different==== | ====Other Properties of Tn''10'': The Flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10L IS''10''] are Functionally Different==== | ||
| Line 110: | Line 110: | ||
====Other Properties of Tn''10'': Tn10 Transposition frequency as a function of length (Transposition from a λ prophage)==== | ====Other Properties of Tn''10'': Tn10 Transposition frequency as a function of length (Transposition from a λ prophage)==== | ||
| − | The frequency of [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] transposition was observed to decrease exponentially with increasing length (distance between the two flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10L IS''10'']) (Morisato et al 1983 PMID: 6299577) as had previously been observed for Tn''9'' (Chandler et al. 1982 PMID: 6281439). This was measured with a mating out assay using an F’lacpro plasmid as a target and a series of isogenic [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] elements carried in a phage hisG::[https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] λ prophage whose length was varied largely by deletion of parts of the internal DNA segment to generate derivative of between full-length (9147bp) and ~4500bp. It was stated that similar result was obtained using a hop assay. In a first set of experiments, [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] derivatives with active transposase were used. In this case, transposition was said to be “self-driven” (Fig. Tn10. | + | The frequency of [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] transposition was observed to decrease exponentially with increasing length (distance between the two flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10L IS''10'']) (Morisato et al 1983 PMID: 6299577) as had previously been observed for Tn''9'' (Chandler et al. 1982 PMID: 6281439). This was measured with a mating out assay using an F’lacpro plasmid as a target and a series of isogenic [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] elements carried in a phage hisG::[https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] λ prophage whose length was varied largely by deletion of parts of the internal DNA segment to generate derivative of between full-length (9147bp) and ~4500bp. It was stated that similar result was obtained using a hop assay. In a first set of experiments, [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] derivatives with active transposase were used. In this case, transposition was said to be “self-driven” ([[:File:FigTn10-5.png|Fig. Tn10.5 '''A''']]). Transposition frequencies decreased by a factor of 40% for every 1kb increase in transposon length. Moreover, a curve with a similar slope but with higher absolute frequency values was observed if [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] transposase was supplied ''in trans'' from two different multicopy plasmids to [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] derivatives with non-functional transposase ([[:File:FigTn10-5.png|Fig. Tn10.5 '''A''']]). Plasmid pNK290 contains an [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''R] with a regulatory mutation which increases [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] transpositon 100 fold while in pNK432, the transposase gene is driven by the histidine operon promoter. The dotted arrows show derivatives which are more active or less active than expected for unknown reasons. |
[[File:FigTn10-5.png|center|thumb|700x700px|'''Fig. Tn10.5. Length dependence of Tn10 transposition obtained using a mating-out assay.''' '''A)''' '''Transposition from a λ prophage.''' “Self-driven” shows the frequencies of [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] derivatives which provide their own transposase. Experiments with pNK290 and pNK432 include both self-driven and defective [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] derivatives. The multicopy plasmid pNK290 carries [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''R] with an upstream regulatory mutation increasing [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] transposition 100x. The multicopy plasmid pNK432 carries an [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''R] in which the transposase expression is driven by the strong ''his'' operon promoter. The scale at the bottom shows transposon length in kilobases <ref><nowiki><pubmed> 6299577</pubmed></nowiki></ref>. '''B) Transposition from a small plasmid. Top:''' Structure of the 14.8 kb plasmid pNK615. Each of the four TnJO termini consists of the outermost 70 bp of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10L IS''10''] ('''right'''). The two pairs of termini bracket 4.0 kb KanR and 5.0 kb LacZ segments, respectively. The HisD and AmpR-Ori segments are each 2.9 kb. Left hand column shows the restriction sites used to add of remove DNA; the second column indicates insertions and deletions in the segment and their size; the third, fourth and fifth columns show the relative transposition frequency of each segment as a function of the size (insertion or deletion of DNA) <ref name=":0"><nowiki><pubmed> 2998922</pubmed></nowiki></ref>.]] | [[File:FigTn10-5.png|center|thumb|700x700px|'''Fig. Tn10.5. Length dependence of Tn10 transposition obtained using a mating-out assay.''' '''A)''' '''Transposition from a λ prophage.''' “Self-driven” shows the frequencies of [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] derivatives which provide their own transposase. Experiments with pNK290 and pNK432 include both self-driven and defective [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] derivatives. The multicopy plasmid pNK290 carries [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''R] with an upstream regulatory mutation increasing [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] transposition 100x. The multicopy plasmid pNK432 carries an [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''R] in which the transposase expression is driven by the strong ''his'' operon promoter. The scale at the bottom shows transposon length in kilobases <ref><nowiki><pubmed> 6299577</pubmed></nowiki></ref>. '''B) Transposition from a small plasmid. Top:''' Structure of the 14.8 kb plasmid pNK615. Each of the four TnJO termini consists of the outermost 70 bp of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10L IS''10''] ('''right'''). The two pairs of termini bracket 4.0 kb KanR and 5.0 kb LacZ segments, respectively. The HisD and AmpR-Ori segments are each 2.9 kb. Left hand column shows the restriction sites used to add of remove DNA; the second column indicates insertions and deletions in the segment and their size; the third, fourth and fifth columns show the relative transposition frequency of each segment as a function of the size (insertion or deletion of DNA) <ref name=":0"><nowiki><pubmed> 2998922</pubmed></nowiki></ref>.]] | ||
| Line 117: | Line 117: | ||
<br /> | <br /> | ||
====Other Properties of Tn''10'': Lack of Length Dependence Using a Plasmid Donor==== | ====Other Properties of Tn''10'': Lack of Length Dependence Using a Plasmid Donor==== | ||
| − | A second set of experiments <ref name=":0" /> indicated that length dependent transposition does not occur if circular plasmids are used as donors. (WAS THIS PREDICTED BY US???). Using a plasmid, pNK615, which carries a number of appropriately oriented [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10L IS''10''] ends (Fig. Tn10. | + | A second set of experiments <ref name=":0" /> indicated that length dependent transposition does not occur if circular plasmids are used as donors. (WAS THIS PREDICTED BY US???). Using a plasmid, pNK615, which carries a number of appropriately oriented [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10L IS''10''] ends ([[:File:FigTn10-5.png|Fig. Tn10.5 '''B''' '''Top''']]) as a donor in a mating out assay. The frequencies of transposition of each segment cannot be directly compared since factors such as impinging transcription and DAM [[wikipedia:Methylation|methylation]] in the '''IR''' can profoundly influence transposition. To overcome these limitation, various insertions or deletions were introduced into one of three positions creating large variations in length ([[:File:FigTn10-5.png|Fig. Tn10.5 '''B''' '''Bottom''']]) and transposition frequencies were normalized to the non-modified pNK615 segments. Only small differences were noted. |
This is perhaps not unexpected since presumably, the transposition frequency would be determined by the shorted distance between the appropriately oriented ends rather that solely the distance in the transposon (see Chandler et al., <ref><nowiki><pubmed>6281439</pubmed></nowiki></ref> for discussion) | This is perhaps not unexpected since presumably, the transposition frequency would be determined by the shorted distance between the appropriately oriented ends rather that solely the distance in the transposon (see Chandler et al., <ref><nowiki><pubmed>6281439</pubmed></nowiki></ref> for discussion) | ||
| Line 123: | Line 123: | ||
====Origin==== | ====Origin==== | ||
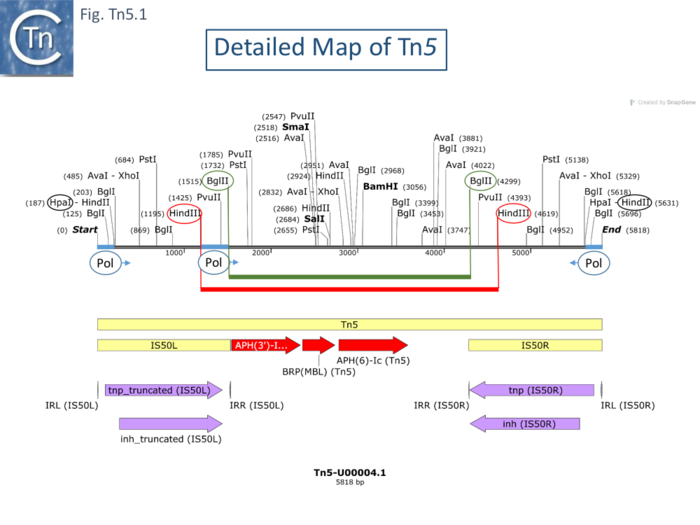
| − | [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] (Fig. Tn5.1) was isolated by transposition to [[wikipedia:Lambda_phage|bacteriophage λ]] from the ''[[wikipedia:Klebsiella|Klebsiella]]'' resistance plasmid JR67 which had been transferred to ''[[wikipedia:Escherichia_coli|E. coli]]'' <ref name=":8" />. It is structurally similar to [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] with two long inverted flanking repeats of about 1.45kb. These were subsequently called [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50'']. Like [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''], it transposes at relatively high frequencies, at many different sites and is mutagenic, generating polar mutations in [[wikipedia:Operon|operons]] on insertion. | + | [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] ([[:File:FigTn5-1.png|Fig. Tn5.1]]) was isolated by transposition to [[wikipedia:Lambda_phage|bacteriophage λ]] from the ''[[wikipedia:Klebsiella|Klebsiella]]'' resistance plasmid JR67 which had been transferred to ''[[wikipedia:Escherichia_coli|E. coli]]'' <ref name=":8" />. It is structurally similar to [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] with two long inverted flanking repeats of about 1.45kb. These were subsequently called [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50'']. Like [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''], it transposes at relatively high frequencies, at many different sites and is mutagenic, generating polar mutations in [[wikipedia:Operon|operons]] on insertion. |
<br /> | <br /> | ||
[[File:FigTn5-1.png|center|thumb|700x700px|'''Fig. Tn5.1. Detailed map of Tn''5'' element.''']] | [[File:FigTn5-1.png|center|thumb|700x700px|'''Fig. Tn5.1. Detailed map of Tn''5'' element.''']] | ||
| Line 149: | Line 149: | ||
====The Inverted Repeats of [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] are Functionally Different.==== | ====The Inverted Repeats of [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] are Functionally Different.==== | ||
| − | Like those of [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''], the flanking [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] repeats are functionally different <ref name=":22"><nowiki><pubmed>6244898</pubmed></nowiki></ref>. An extensive restriction endonuclease map (Fig. Comp.7) <ref name=":14" /> was used to design a library of [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] mutants. This localized the ''aph'' gene proximal to the inside end ('''IE''') of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS50L]. Insertions at the left ''Bgl''II site were found to severely reduce neomycin resistance levels <ref name=":14" /> suggesting that ''aph'' expression could be driven from [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L]. This was confirmed by the observation that inversion of a ''Bgl''II fragment resulted in loss of resistance while resistance is retained on inversion of a ''Hind''III fragment (Fig. Tn5.1) <ref name=":22" />. | + | Like those of [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''], the flanking [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] repeats are functionally different <ref name=":22"><nowiki><pubmed>6244898</pubmed></nowiki></ref>. An extensive restriction endonuclease map (F[[:File:Fig-Comp1.png|ig. Comp.7]]) <ref name=":14" /> was used to design a library of [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] mutants. This localized the ''aph'' gene proximal to the inside end ('''IE''') of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS50L]. Insertions at the left ''Bgl''II site were found to severely reduce neomycin resistance levels <ref name=":14" /> suggesting that ''aph'' expression could be driven from [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L]. This was confirmed by the observation that inversion of a ''Bgl''II fragment resulted in loss of resistance while resistance is retained on inversion of a ''Hind''III fragment ([[:File:FigTn5-1.png|Fig. Tn5.1]]) <ref name=":22" />. |
| − | RNA polymerase binding sites were located using gel mobility shift assays: binding occurred in DNA fragments at both outside ends ('''OE''') and a third weak binding site was located close to IE of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] but not in this location in [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R] (Fig. Tn5.1). The direction of transcription was determined ''in vitro'' using different size fragments and determining the length of the run-off transcripts. It was found to start at '''OE''' and continue through the IS. Surprisingly, two distinct transcripts were identified differing at their 5’ end by about 10 nucleotides. | + | RNA polymerase binding sites were located using gel mobility shift assays: binding occurred in DNA fragments at both outside ends ('''OE''') and a third weak binding site was located close to IE of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] but not in this location in [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R] ([[:File:FigTn5-1.png|Fig. Tn5.1]]). The direction of transcription was determined ''in vitro'' using different size fragments and determining the length of the run-off transcripts. It was found to start at '''OE''' and continue through the IS. Surprisingly, two distinct transcripts were identified differing at their 5’ end by about 10 nucleotides. |
| − | Five distinct [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5'']-specific translation products were identified using a minicell system (of 58K, 54K, 53K, 49K and 26K). A major product had a mass, 26Kd, consistent with that expected for the [[wikipedia:Neomycin|neomycin phophotransferase]]. The sum of the other 4 species was clearly significantly larger than the coding capacity of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''] elements. The smallest of these species, 49Kd, had previously been shown to be encoded by [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] <ref><nowiki><pubmed>390162</pubmed></nowiki></ref>. An insertion into the ''Hin''dIII site of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] resulted in the loss of the 53Kd and 49Kd species whereas an insertion into the ''Hin''dIII site in [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R] resulted in the loss of the 58K and 54K species <ref name=":22" />. The assignment of the species to [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] or [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R] was confirmed by deletion of one or other of the IS. Concomitant with the loss of the 4 protein species, insertions in the ''Hin''dIII sites of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R] both gave rise to species of 42K and 38K, fusion protein products resulting from the inserted DNA. These results imply that the difference in size of the two proteins from one IS derives from a difference in their N-terminal ends whereas the difference in size of the proteins between the two IS results from their C-terminal ends (Fig. Tn5.1). To further localize the open reading frames, a [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] derivative deleted for both ends at the ''Hpa''I-''Hind''II site does not produce the 58K and 53K species, since the RNA pol binding site is removed, and produces reduced amounts of the 54K and 49K proteins (probably due to impinging transcription from the surrounding sequence context). | + | Five distinct [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5'']-specific translation products were identified using a minicell system (of 58K, 54K, 53K, 49K and 26K). A major product had a mass, 26Kd, consistent with that expected for the [[wikipedia:Neomycin|neomycin phophotransferase]]. The sum of the other 4 species was clearly significantly larger than the coding capacity of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''] elements. The smallest of these species, 49Kd, had previously been shown to be encoded by [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] <ref><nowiki><pubmed>390162</pubmed></nowiki></ref>. An insertion into the ''Hin''dIII site of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] resulted in the loss of the 53Kd and 49Kd species whereas an insertion into the ''Hin''dIII site in [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R] resulted in the loss of the 58K and 54K species <ref name=":22" />. The assignment of the species to [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] or [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R] was confirmed by deletion of one or other of the IS. Concomitant with the loss of the 4 protein species, insertions in the ''Hin''dIII sites of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R] both gave rise to species of 42K and 38K, fusion protein products resulting from the inserted DNA. These results imply that the difference in size of the two proteins from one IS derives from a difference in their N-terminal ends whereas the difference in size of the proteins between the two IS results from their C-terminal ends ([[:File:FigTn5-1.png|Fig. Tn5.1]]). To further localize the open reading frames, a [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] derivative deleted for both ends at the ''Hpa''I-''Hind''II site does not produce the 58K and 53K species, since the RNA pol binding site is removed, and produces reduced amounts of the 54K and 49K proteins (probably due to impinging transcription from the surrounding sequence context). |
To determine whether these differences between [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R] are reflected in their abilities to drive [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn5] transposition, a number of mutations were introduced into either [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] or [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R]. Ablation of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R] consistently had a severe effect on transposition of the [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] derivative whereas ablation of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] had only a small effect. [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R] makes a major contribution to [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] transposition while [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] is necessary for expression of the [[wikipedia:Neomycin|neomycin resistance gene]]. | To determine whether these differences between [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R] are reflected in their abilities to drive [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn5] transposition, a number of mutations were introduced into either [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] or [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R]. Ablation of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R] consistently had a severe effect on transposition of the [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] derivative whereas ablation of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] had only a small effect. [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''R] makes a major contribution to [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] transposition while [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''L] is necessary for expression of the [[wikipedia:Neomycin|neomycin resistance gene]]. | ||
| Line 170: | Line 170: | ||
====Origin==== | ====Origin==== | ||
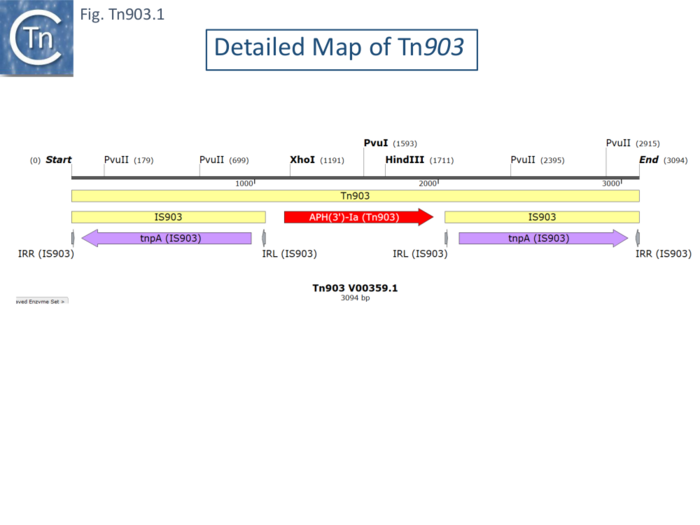
| − | [https://tncentral.ncc.unesp.br/report/te/Tn903-V00359.1 Tn''903''] was originally isolated from plasmid pML21 <ref><nowiki><pubmed>770430</pubmed></nowiki></ref> (a hybrid plasmid consisting of pVH51 and the Km<sup>R</sup> fragment of plasmid pSC105 inserted at the ''Eco''RI restriction site of mini-ColE1), by transposition onto the phage fd <ref name=":23"><nowiki><pubmed>344143</pubmed></nowiki></ref> and shown to transpose to multiple sites in other replicons. [https://tncentral.ncc.unesp.br/report/te/Tn903-V00359.1 Tn''903''] transposition activity is low, and it has previously been used as a stable selective marker on many plasmids used as cloning vehicles. It is carried by plasmid R6(-5) <ref><nowiki><pubmed>4580676</pubmed></nowiki></ref>. It was shown to comprise a central region of about 1kb carrying a [[wikipedia:Kanamycin_A|kanamycin resistance]] gene flanked by 1050bp inverted repeats <ref name=":23" /> (Fig. Tn903.1) which were called [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] <ref name=":1"><nowiki><pubmed>6261245</pubmed></nowiki></ref>. Insertion results in a target duplication of 9bp but insertion sites did not appear to have any common sequence features <ref name=":11" />. Deletion of an internal restriction fragment from both repeats eliminated [https://tncentral.ncc.unesp.br/report/te/Tn903-V00359.1 Tn''903''] transposition but deletions of the fragment from only one repeat retained transposition activity. Moreover, an isolated [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] copy was shown to undergo independent transposition <ref name=":1" />. The complete nucleotide sequence was obtained by Oka et al <ref><nowiki><pubmed>6270337</pubmed></nowiki></ref> who identified the [[wikipedia:Kanamycin_A|kanamycin resistance]] (APH) gene and its product in a minicell system. | + | [https://tncentral.ncc.unesp.br/report/te/Tn903-V00359.1 Tn''903''] was originally isolated from plasmid pML21 <ref><nowiki><pubmed>770430</pubmed></nowiki></ref> (a hybrid plasmid consisting of pVH51 and the Km<sup>R</sup> fragment of plasmid pSC105 inserted at the ''Eco''RI restriction site of mini-ColE1), by transposition onto the phage fd <ref name=":23"><nowiki><pubmed>344143</pubmed></nowiki></ref> and shown to transpose to multiple sites in other replicons. [https://tncentral.ncc.unesp.br/report/te/Tn903-V00359.1 Tn''903''] transposition activity is low, and it has previously been used as a stable selective marker on many plasmids used as cloning vehicles. It is carried by plasmid R6(-5) <ref><nowiki><pubmed>4580676</pubmed></nowiki></ref>. It was shown to comprise a central region of about 1kb carrying a [[wikipedia:Kanamycin_A|kanamycin resistance]] gene flanked by 1050bp inverted repeats <ref name=":23" /> ([[:File:FigTn903-1.png|Fig. Tn903.1]]) which were called [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] <ref name=":1"><nowiki><pubmed>6261245</pubmed></nowiki></ref>. Insertion results in a target duplication of 9bp but insertion sites did not appear to have any common sequence features <ref name=":11" />. Deletion of an internal restriction fragment from both repeats eliminated [https://tncentral.ncc.unesp.br/report/te/Tn903-V00359.1 Tn''903''] transposition but deletions of the fragment from only one repeat retained transposition activity. Moreover, an isolated [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] copy was shown to undergo independent transposition <ref name=":1" />. The complete nucleotide sequence was obtained by Oka et al <ref><nowiki><pubmed>6270337</pubmed></nowiki></ref> who identified the [[wikipedia:Kanamycin_A|kanamycin resistance]] (APH) gene and its product in a minicell system. |
[[File:FigTn903-1.png|center|thumb|700x700px|'''Fig. Tn903.1. Detailed map of Tn''903.''''']] | [[File:FigTn903-1.png|center|thumb|700x700px|'''Fig. Tn903.1. Detailed map of Tn''903.''''']] | ||
| Line 179: | Line 179: | ||
====Cointegrate formation?==== | ====Cointegrate formation?==== | ||
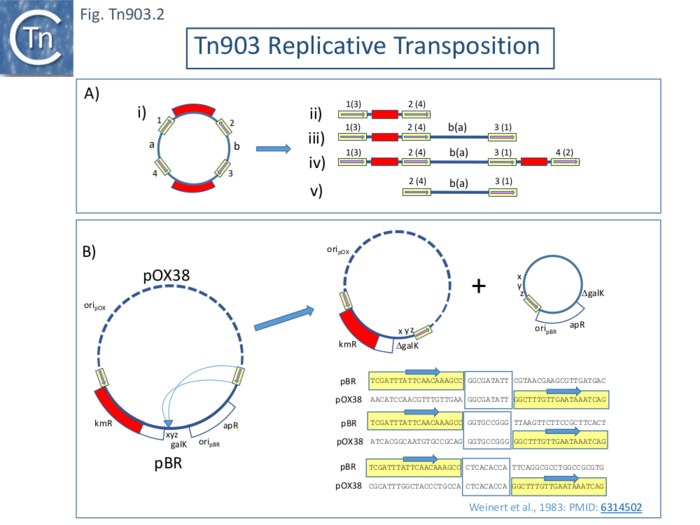
| − | Replicative transposition, is often evidenced by the formation of cointegrates. However, it should be underlined that identification of cointegrate structures does not necessarily indicate replicative transposition since they can be formed by direct transposition from a dimeric donor plasmid (Fig. Tn903. | + | Replicative transposition, is often evidenced by the formation of cointegrates. However, it should be underlined that identification of cointegrate structures does not necessarily indicate replicative transposition since they can be formed by direct transposition from a dimeric donor plasmid ([[:File:FigTn903-2.png|Fig. Tn903.2 '''A''']] where ii is a direct transposition and v an inverse transposition while both iii and iv resemble replicative cointegrates but in fact are formed by direct transposition). Plasmid dimerization can occur even in the absence of the ''recA'' function <ref name=":2"><nowiki><pubmed>7029308</pubmed></nowiki></ref>. The monomer-dimer plasmid status might also be impacted by the recombination status of the host although plasmid dimerization can occur even in the absence of the ''recA'' function <ref name=":2" />. This had been noted in the case of [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] <ref><nowiki><pubmed>6292435</pubmed></nowiki></ref> where cointegrate formation was found to be dependent on the ''recA'' function <ref><nowiki><pubmed>6289304</pubmed></nowiki></ref> and was ascribed to direct transposition from plasmid dimers <ref><nowiki><pubmed>6298776</pubmed></nowiki></ref>. |
====Replicative Intramolecular Transposition==== | ====Replicative Intramolecular Transposition==== | ||
| − | To address the question of replicative transposition of [https://tncentral.ncc.unesp.br/report/te/Tn903-V00359.1 Tn''903''], Weinert et al <ref name=":24" /><ref name=":25" /> investigated intramolecular transposition using a cloned neighboring ''galK'' gene copy next to [https://tncentral.ncc.unesp.br/report/te/Tn903-V00359.1 Tn''903''] in a pBR322-derivative plasmid. High [[wikipedia:Galactokinase|galactokinase]] (GalK) expression is lethal in an ''[[wikipedia:Escherichia_coli|E. coli]]'' ''galE''-''T''- strain in the presence of [[wikipedia:Galactose|galactose]]. Selection of galactose resistance selects for mutation/deletion/rearrangement of ''galK''. Deletions using a highly active [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] retained the IS and eliminated DNA from through into and through ''galK'' as would be expected to result from an intramolecular replicative cointegrate formation with the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] repeat joined to a new target sequence (see: [[Transposons families/Tn3 family|Tn''3'' family]] and [[IS Families/IS6 family|IS''6'' family]]). It was further shown that deletion require both [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] ends. Using a cointegrate formed between the ''galK'' pBR322 plasmid and pOX38, it was possible to identify both parts of the deletion product, since each carries a replicative origin, using their different drug resistances (Fig. Tn903. | + | To address the question of replicative transposition of [https://tncentral.ncc.unesp.br/report/te/Tn903-V00359.1 Tn''903''], Weinert et al <ref name=":24" /><ref name=":25" /> investigated intramolecular transposition using a cloned neighboring ''galK'' gene copy next to [https://tncentral.ncc.unesp.br/report/te/Tn903-V00359.1 Tn''903''] in a pBR322-derivative plasmid. High [[wikipedia:Galactokinase|galactokinase]] (GalK) expression is lethal in an ''[[wikipedia:Escherichia_coli|E. coli]]'' ''galE''-''T''- strain in the presence of [[wikipedia:Galactose|galactose]]. Selection of galactose resistance selects for mutation/deletion/rearrangement of ''galK''. Deletions using a highly active [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] retained the IS and eliminated DNA from through into and through ''galK'' as would be expected to result from an intramolecular replicative cointegrate formation with the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] repeat joined to a new target sequence (see: [[Transposons families/Tn3 family|Tn''3'' family]] and [[IS Families/IS6 family|IS''6'' family]]). It was further shown that deletion require both [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] ends. Using a cointegrate formed between the ''galK'' pBR322 plasmid and pOX38, it was possible to identify both parts of the deletion product, since each carries a replicative origin, using their different drug resistances ([[:File:FigTn903-2.png|Fig. Tn903.2 '''B''']]). Additionally, intramolecular replicative transposition should generate a direct target repeat at the insertion point which would abut each new junction in each deletion product. The authors identified such paired 9bp sequences as expected providing strong evidence for a replicative transposition process ([[:File:FigTn903-2.png|Fig. Tn903.2 '''B''']]). |
[[File:FigTn903-2.png|center|thumb|700x700px|'''Fig. Tn903.2. Tn903 Replicative Transpostion.''']] | [[File:FigTn903-2.png|center|thumb|700x700px|'''Fig. Tn903.2. Tn903 Replicative Transpostion.''']] | ||
<br /> | <br /> | ||
| Line 201: | Line 201: | ||
====Origin==== | ====Origin==== | ||
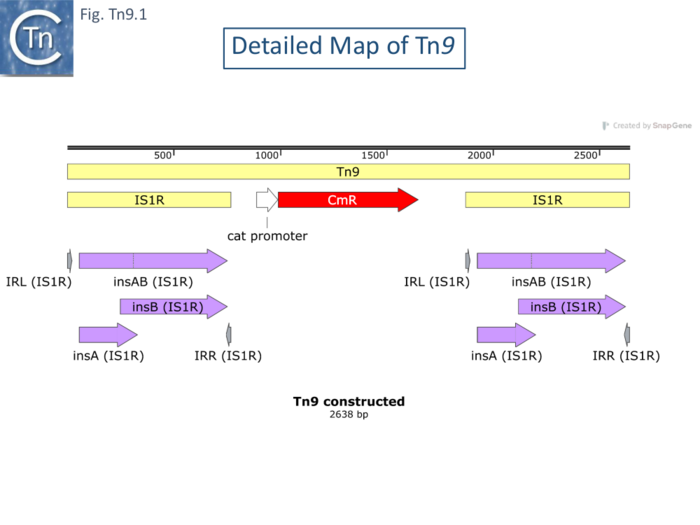
| − | Like [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] and [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''], Tn''9'' (Fig. Tn9.1) was among the first compound transposons to be identified. Following the demonstration that [[wikipedia:P1_phage|phage P1]] could acquire [[wikipedia:Chloramphenicol|chloramphenicol]] resistance from a drug resistance plasmid <ref name=":5" />, and its subsequent acquisition by phage<ref name=":6" /> <ref name=":7" /> isolated λ cam phage from a P1cam lysogen by thermal induction of λcI857. They showed that λcam phage were denser than the parental phage in CsCl gradients – indicating that they contained a DNA insertion and localized this by heteroduplex analysis. Acquisition appeared independent of the host ''rec'' and phage ''red'' recombination systems. They observed that insertions of identical size could occur at various places in the phage. In a series of articles in DNA Insertion Elements,Plasmids and Episomes (1977 ISBN0-87969-118-2), Rosner and Gottesman, Chow and Bukhari, MacHattie and Jakowski, and Yun and Vapnek all showed that similar sized [[wikipedia:Chloramphenicol|chloramphenicol]] resistant insertions in phage P1 and Mu or phage λ basically using heteroduplex analysis. MacHattie and Jakowski used heteroduplexes with λ phage carrying insertions of IS''1'' to demonstrate that the cam insertions carried flanking directly repeated [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'' copies]. Using restriction analysis and Southern hybridization of [[wikipedia:Chloramphenicol|chloramphenicol]] resistant insertions in [[wikipedia:P1_phage|phage P1]] and [[wikipedia:Bacteriophage_Mu|phage Mu]] <ref><nowiki><pubmed>365686</pubmed></nowiki></ref>and concluded that Tn''9'' was flanked by two directly repeated copies of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']. | + | Like [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] and [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''], Tn''9'' ([[:File:FigTn9-1.png|Fig. Tn9.1]]) was among the first compound transposons to be identified. Following the demonstration that [[wikipedia:P1_phage|phage P1]] could acquire [[wikipedia:Chloramphenicol|chloramphenicol]] resistance from a drug resistance plasmid <ref name=":5" />, and its subsequent acquisition by phage<ref name=":6" /> <ref name=":7" /> isolated λ cam phage from a P1cam lysogen by thermal induction of λcI857. They showed that λcam phage were denser than the parental phage in CsCl gradients – indicating that they contained a DNA insertion and localized this by heteroduplex analysis. Acquisition appeared independent of the host ''rec'' and phage ''red'' recombination systems. They observed that insertions of identical size could occur at various places in the phage. In a series of articles in DNA Insertion Elements,Plasmids and Episomes (1977 ISBN0-87969-118-2), Rosner and Gottesman, Chow and Bukhari, MacHattie and Jakowski, and Yun and Vapnek all showed that similar sized [[wikipedia:Chloramphenicol|chloramphenicol]] resistant insertions in phage P1 and Mu or phage λ basically using heteroduplex analysis. MacHattie and Jakowski used heteroduplexes with λ phage carrying insertions of IS''1'' to demonstrate that the cam insertions carried flanking directly repeated [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'' copies]. Using restriction analysis and Southern hybridization of [[wikipedia:Chloramphenicol|chloramphenicol]] resistant insertions in [[wikipedia:P1_phage|phage P1]] and [[wikipedia:Bacteriophage_Mu|phage Mu]] <ref><nowiki><pubmed>365686</pubmed></nowiki></ref>and concluded that Tn''9'' was flanked by two directly repeated copies of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']. |
[[File:FigTn9-1.png|center|thumb|700x700px|'''Fig. Tn9.1. Detailed map of Tn9.''']] | [[File:FigTn9-1.png|center|thumb|700x700px|'''Fig. Tn9.1. Detailed map of Tn9.''']] | ||
Kondo and Mitsuhashi <ref name=":5" /> observed that 5-20% of P1cam phage lose Cam<sup>R</sup> on growth and this was accompanied by a decrease in density indicating loss of DNA. It was also noted that Cam<sup>S</sup> phage retained part of the original insertion a length of DNA equivalent to the length of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']. Similar instability was observed by MacHattie and Jakowski and by Gottesman and Rosner (1977 ISBN0-87969-118-2) using phage λ cam where a single plate lysate could include 10 - 70% Cam<sup>S</sup> phage. Presumably these deletions occur by reciprocal recombination between the flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']. | Kondo and Mitsuhashi <ref name=":5" /> observed that 5-20% of P1cam phage lose Cam<sup>R</sup> on growth and this was accompanied by a decrease in density indicating loss of DNA. It was also noted that Cam<sup>S</sup> phage retained part of the original insertion a length of DNA equivalent to the length of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']. Similar instability was observed by MacHattie and Jakowski and by Gottesman and Rosner (1977 ISBN0-87969-118-2) using phage λ cam where a single plate lysate could include 10 - 70% Cam<sup>S</sup> phage. Presumably these deletions occur by reciprocal recombination between the flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']. | ||
| Line 211: | Line 211: | ||
[[File:FigTn9-2.png|center|thumb|700x700px|'''Fig. Tn9.2. Amplification of Tn9 shown by agarose gel electrophoresis'''. Pattern of fragments obtained by digestion of pLClI0, an R100.1 RTF plasmid carrying an Tn''9'' insertion and RTF with ''Eco''RI. Electrophoresis was on l% agarose slab gels: Lanes 1 and 2: pLCll0 isolated from a recA- ''[[wikipedia:Escherichia_coli|E. coli]]'' strain in the absence and presence respectively of [[wikipedia:Chloramphenicol|chloramphenicol]]. Lanes 3 and 4; pLCll0 isolated from a recA + strain in the absence and presence respectively of [[wikipedia:Chloramphenicol|chloramphenicol]]. Lane 5: Parental RTF DNA. The notation used for the RTF fragments is shown on the left of this figure, '''b)''' Autoradiograph of the gel shown in '''a)''' following denaturation, transfer to a nitrocellulose filter and hybridization with 3~P-labeled 2::IS1 DNA. The RTF fragment which has previously been shown to carry [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'' homologous] material is fragment A'. '''c)''' R100.1 DNA isolated from ceils grown in the presence (lane 1) and absence (lane 2) of [[wikipedia:Chloramphenicol|chloramphenicol]]. The notation used for the R100.1 fragments is shown on the right of this figure. The position of fragment AH, expected to arise from tandem duplication (Tanaka et al., 1976) or circularisation (Chandler et al., 1977b) of the r-det is indicated by the arrow]] | [[File:FigTn9-2.png|center|thumb|700x700px|'''Fig. Tn9.2. Amplification of Tn9 shown by agarose gel electrophoresis'''. Pattern of fragments obtained by digestion of pLClI0, an R100.1 RTF plasmid carrying an Tn''9'' insertion and RTF with ''Eco''RI. Electrophoresis was on l% agarose slab gels: Lanes 1 and 2: pLCll0 isolated from a recA- ''[[wikipedia:Escherichia_coli|E. coli]]'' strain in the absence and presence respectively of [[wikipedia:Chloramphenicol|chloramphenicol]]. Lanes 3 and 4; pLCll0 isolated from a recA + strain in the absence and presence respectively of [[wikipedia:Chloramphenicol|chloramphenicol]]. Lane 5: Parental RTF DNA. The notation used for the RTF fragments is shown on the left of this figure, '''b)''' Autoradiograph of the gel shown in '''a)''' following denaturation, transfer to a nitrocellulose filter and hybridization with 3~P-labeled 2::IS1 DNA. The RTF fragment which has previously been shown to carry [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'' homologous] material is fragment A'. '''c)''' R100.1 DNA isolated from ceils grown in the presence (lane 1) and absence (lane 2) of [[wikipedia:Chloramphenicol|chloramphenicol]]. The notation used for the R100.1 fragments is shown on the right of this figure. The position of fragment AH, expected to arise from tandem duplication (Tanaka et al., 1976) or circularisation (Chandler et al., 1977b) of the r-det is indicated by the arrow]] | ||
| − | Tn9 was observed to form tandem dimers when harbored by small [[wikipedia:Lambda_phage|phage λ]] derivatives with smaller genomes capable of accommodating larger pieces of inserted DNA (MacHattie and Jakowski 1977 ISBN0-87969-118-2). This phenomenon was also observed when Tn''9'' was inserted into the resistance transfer factor (a derivative of R100.1 which lacks the major drug resistance gene cluster <ref><nowiki><pubmed>393954</pubmed></nowiki></ref>; and the culture was grown with consecutive increases in the concentration of [[wikipedia:Chloramphenicol|chloramphenicol]] in the medium up to 1mg/ml. Tn''9'' contains a single ''Eco''RI site in the Cm gene and ''Eco''RI digestion generated a fragment specific for Tn''9'' which can be detected simply in agarose gels (Fig. Tn9. | + | Tn9 was observed to form tandem dimers when harbored by small [[wikipedia:Lambda_phage|phage λ]] derivatives with smaller genomes capable of accommodating larger pieces of inserted DNA (MacHattie and Jakowski 1977 ISBN0-87969-118-2). This phenomenon was also observed when Tn''9'' was inserted into the resistance transfer factor (a derivative of R100.1 which lacks the major drug resistance gene cluster <ref><nowiki><pubmed>393954</pubmed></nowiki></ref>; and the culture was grown with consecutive increases in the concentration of [[wikipedia:Chloramphenicol|chloramphenicol]] in the medium up to 1mg/ml. Tn''9'' contains a single ''Eco''RI site in the Cm gene and ''Eco''RI digestion generated a fragment specific for Tn''9'' which can be detected simply in agarose gels ([[:File:FigTn9-2.png|Fig. Tn9.2 '''A''']] '''lane 4''') or following a Southern blot using an [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']-specific probe ([[:File:FigTn9-2.png|Fig. Tn9.2 '''B''']] '''lane 4'''). |
====Two Transposition Pathways: Direct Transposition and Tn''9''-mediated Cointegrate Formation?==== | ====Two Transposition Pathways: Direct Transposition and Tn''9''-mediated Cointegrate Formation?==== | ||
It was deduced by studying the frequency of transposition and cointegrate formation and the stability the transposition products that Tn''9'' transposition generates two types of mutually exclusive, end-products: either direct insertion into a recipient replicon (transposition), or fusion between donor and recipient replicons (cointegrate formation). This conclusion is based on the observations that, while Tn''9''-mediated cointegrates are very stable, they are formed at a rate lower than the transposition frequency. The ability of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] to promote these events is therefore also reflected in the behavior of Tn''9''. | It was deduced by studying the frequency of transposition and cointegrate formation and the stability the transposition products that Tn''9'' transposition generates two types of mutually exclusive, end-products: either direct insertion into a recipient replicon (transposition), or fusion between donor and recipient replicons (cointegrate formation). This conclusion is based on the observations that, while Tn''9''-mediated cointegrates are very stable, they are formed at a rate lower than the transposition frequency. The ability of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] to promote these events is therefore also reflected in the behavior of Tn''9''. | ||
| Line 217: | Line 217: | ||
The capacity of Tn''9'' and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] to generate cointegrates and to undergo transposition were measured in three ways: transposition from a λ::Tn''9'' prophage to an F-derived plasmid pOX38; cointegrate formation between pOX38::Tn''9'' and the non-conjugative plasmid, pBR322; and both cointegrate formation and transposition between the plasmid pBR322::Tn''9'' and pOX38 <ref><nowiki><pubmed>6281440</pubmed></nowiki></ref>. | The capacity of Tn''9'' and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] to generate cointegrates and to undergo transposition were measured in three ways: transposition from a λ::Tn''9'' prophage to an F-derived plasmid pOX38; cointegrate formation between pOX38::Tn''9'' and the non-conjugative plasmid, pBR322; and both cointegrate formation and transposition between the plasmid pBR322::Tn''9'' and pOX38 <ref><nowiki><pubmed>6281440</pubmed></nowiki></ref>. | ||
| − | Three types of cointegrates were observed: replicon fusions using one or the other flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] and examples where the entire Tn''9'' had been used (Fig.Tn9.3). These structures were examined using restriction mapping of 40 individual clones using restriction mapping together with electron microscopy of 8 examples for confirmation. Of the 33 which used [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1A IS''1''A], 20 were simple fusions using [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1A IS''1''A] (Fig.Tn9. | + | Three types of cointegrates were observed: replicon fusions using one or the other flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] and examples where the entire Tn''9'' had been used ([[:File:FigTn9-3.png|Fig.Tn9.3]]). These structures were examined using restriction mapping of 40 individual clones using restriction mapping together with electron microscopy of 8 examples for confirmation. Of the 33 which used [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1A IS''1''A], 20 were simple fusions using [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1A IS''1''A] ([[:File:FigTn9-3.png|Fig.Tn9.3 '''B''']]), 10 contained tandem copies of the pBR322 ([[:File:FigTn9-3.png|Fig.Tn9.3 '''C''']]) and 3 used [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1A IS''1''A] but had multiple tandem pBR322 and Tn''9'' copies. Of the 4 which used [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1B IS''1''B], 3 were simple fusions ([[:File:FigTn9-3.png|Fig.Tn9.3 '''D''']]) and 1 carried a tandem pBR322 dimer ([[:File:FigTn9-3.png|Fig.Tn9.3 '''E''']]). Three examples had used the full length Tn''9''. Two were simple replicon fusions ([[:File:FigTn9-3.png|Fig.Tn9.3 '''F''']]) and one carried a pBR322 Tn''9'' dimer. |
Subsequently, is was shown by identifying a number of transposition intermediates that [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] can transpose in a number of different ways including direct transposition and cointegrate formation <ref><nowiki><pubmed>7489730</pubmed></nowiki></ref> (see: [[IS Families/IS1 family|IS1 Family]]). | Subsequently, is was shown by identifying a number of transposition intermediates that [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] can transpose in a number of different ways including direct transposition and cointegrate formation <ref><nowiki><pubmed>7489730</pubmed></nowiki></ref> (see: [[IS Families/IS1 family|IS1 Family]]). | ||
| Line 232: | Line 232: | ||
Two studies <ref name=":28"><nowiki><pubmed>6309405</pubmed></nowiki></ref><ref name=":29"><nowiki><pubmed>6313938</pubmed></nowiki></ref> explored this notion. The Ohtsubo lab <ref name=":28" /> showed that removal of the ''cam'' promotor in Tn''9'' or the insertion of a phage T7 transcription terminator increased the ability of the downstream [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] to form cointegrates again using pHS1 as a Tn''9'' donor. Moreover, it was demonstrated that ''in vitro'' transcripts of the ''cam'' gene in Tn''9'' read through into the downstream IS. | Two studies <ref name=":28"><nowiki><pubmed>6309405</pubmed></nowiki></ref><ref name=":29"><nowiki><pubmed>6313938</pubmed></nowiki></ref> explored this notion. The Ohtsubo lab <ref name=":28" /> showed that removal of the ''cam'' promotor in Tn''9'' or the insertion of a phage T7 transcription terminator increased the ability of the downstream [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] to form cointegrates again using pHS1 as a Tn''9'' donor. Moreover, it was demonstrated that ''in vitro'' transcripts of the ''cam'' gene in Tn''9'' read through into the downstream IS. | ||
| − | A different approach was used by Chandler and Galas <ref name=":29" /> who came to the same conclusions in a continuing analysis of cointegrate formation. Here, Tn''9'' together with a short segment (132bp) of flanking DNA was cloned in different places and orientations into pBR322 and these were used as donors in a mating out assay using pOX38 as a target. The results showed that the IS1 copy proximal to a promoter exhibits lower activity than the distal copy and that deletion of the divergent promoters P1 and P2 reduces or eliminates this effect (Fig. Tn9.4) | + | A different approach was used by Chandler and Galas <ref name=":29" /> who came to the same conclusions in a continuing analysis of cointegrate formation. Here, Tn''9'' together with a short segment (132bp) of flanking DNA was cloned in different places and orientations into pBR322 and these were used as donors in a mating out assay using pOX38 as a target. The results showed that the IS1 copy proximal to a promoter exhibits lower activity than the distal copy and that deletion of the divergent promoters P1 and P2 reduces or eliminates this effect ([[:File:FigTn9-4.png|Fig. Tn9.4]]) |
| − | [[File:FigTn9-4.png|center|thumb|700x700px|'''Fig. Tn9.4. Impinging Transcription.''' '''A)''' Map of pBR322: top showing cloning sites used ERI, ''Eco''RI; HIII, ''Hin''dIII; EV, ''Eco''RV; BHI, ''Bam''HI; SI, ''Sal''I; and the ''bla'' and ''tet'' genes (red arrows). '''Bottom:''' a more detailed map of the intergenic region showin the position of the known Promoters, P1, P2 and P3 (large blue arrows) together with the direction of transcription (black arrowed lines). '''B)''' and '''C)''' cloning of Tn''9'' into the ''Eco''RI site in opposite orientations. '''D)''' cloning between The ''Bam''HI and ''Sal''I sites. '''E)''', '''F)''' and '''G)''' removal of promoters P1 and P2 by deletion between the ''Hind''III and ''Eco''RV sites. '''H)''' insertion of a transcription terminator Between the ''Hin''dIII and ''Bam''HI sites. Activities were measures in a mating out assay using the F derivative pOX38 as a target plasmid The results expressed as the number of cointegrates formed using [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1A IS''1''A], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1B IS''1''B] and Tn''9'' are shown beneath each figure. Also shown are the number of events occurring by inverse Transposition using the inside ends of the Tn''9'' flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] copies with concomitant loss of the Interstitial ''cm'' gene. Structures were determined in the same way as for those shown in Fig. Tn9.3.]] | + | [[File:FigTn9-4.png|center|thumb|700x700px|'''Fig. Tn9.4. Impinging Transcription.''' '''A)''' Map of pBR322: top showing cloning sites used ERI, ''Eco''RI; HIII, ''Hin''dIII; EV, ''Eco''RV; BHI, ''Bam''HI; SI, ''Sal''I; and the ''bla'' and ''tet'' genes (red arrows). '''Bottom:''' a more detailed map of the intergenic region showin the position of the known Promoters, P1, P2 and P3 (large blue arrows) together with the direction of transcription (black arrowed lines). '''B)''' and '''C)''' cloning of Tn''9'' into the ''Eco''RI site in opposite orientations. '''D)''' cloning between The ''Bam''HI and ''Sal''I sites. '''E)''', '''F)''' and '''G)''' removal of promoters P1 and P2 by deletion between the ''Hind''III and ''Eco''RV sites. '''H)''' insertion of a transcription terminator Between the ''Hin''dIII and ''Bam''HI sites. Activities were measures in a mating out assay using the F derivative pOX38 as a target plasmid The results expressed as the number of cointegrates formed using [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1A IS''1''A], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1B IS''1''B] and Tn''9'' are shown beneath each figure. Also shown are the number of events occurring by inverse Transposition using the inside ends of the Tn''9'' flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] copies with concomitant loss of the Interstitial ''cm'' gene. Structures were determined in the same way as for those shown in [[:File:FigTn9-3.png|Fig. Tn9.3]].]] |
====Tn''9'' Transposition frequency as a function of length: Transposition from a λ prophage==== | ====Tn''9'' Transposition frequency as a function of length: Transposition from a λ prophage==== | ||
| Line 242: | Line 242: | ||
[[File:FigTn9-5.png|center|thumb|700x700px|'''Fig. Tn9.5. Length Dependence of Transposition. Top''': A map of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''-flanked] transposons used. The yellow boxes represent flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'' copies] and their relative orientation. Vertical arrows show the relative positions of ''Eco''RI sites within each transposon. They are taken from the following sources: Tn''9'' <ref><nowiki><pubmed> 390403</pubmed></nowiki></ref>; Tn''2671Δ''202, A72 and 481.1 <ref name=":3" />; Tn''2360'' <ref name=":30" />; Tn''2350Δ''l. M. Clerget (pers. Comm.). '''Bottom''': the proportion of target plasmid (pOX38) which acquired the transposon from a λ prophage after under standard growth conditions as a function of transposon length. Bottom dashed line data using a pOX38 target. Bottom line data using an R100.1 derivative, RTF, which had lost its resistance determinant.]] | [[File:FigTn9-5.png|center|thumb|700x700px|'''Fig. Tn9.5. Length Dependence of Transposition. Top''': A map of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''-flanked] transposons used. The yellow boxes represent flanking [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'' copies] and their relative orientation. Vertical arrows show the relative positions of ''Eco''RI sites within each transposon. They are taken from the following sources: Tn''9'' <ref><nowiki><pubmed> 390403</pubmed></nowiki></ref>; Tn''2671Δ''202, A72 and 481.1 <ref name=":3" />; Tn''2360'' <ref name=":30" />; Tn''2350Δ''l. M. Clerget (pers. Comm.). '''Bottom''': the proportion of target plasmid (pOX38) which acquired the transposon from a λ prophage after under standard growth conditions as a function of transposon length. Bottom dashed line data using a pOX38 target. Bottom line data using an R100.1 derivative, RTF, which had lost its resistance determinant.]] | ||
| − | The results (Fig. Tn9.5) clearly show that with both target plasmids, there is a logarithmic decrease in transposition activity over this size range and is reduced by approximately 50% with each increase of 1kb of DNA. This is somewhat higher than was found with [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn10] (Fig. Tn10.5). One suggestion which has yet to be rigorously demonstrated was that transposition occurs by a progressive tracking mechanism with a constant probability of abortion per unit DNA length. Another would be that the probability of the two [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] outside ends to synapse to form a transpososome simply decreases with increasing distance between the two.<br /> | + | The results ([[:File:FigTn9-5.png|Fig. Tn9.5]]) clearly show that with both target plasmids, there is a logarithmic decrease in transposition activity over this size range and is reduced by approximately 50% with each increase of 1kb of DNA. This is somewhat higher than was found with [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn10] ([[:File:FigTn10-5.png|Fig. Tn10.5]]). One suggestion which has yet to be rigorously demonstrated was that transposition occurs by a progressive tracking mechanism with a constant probability of abortion per unit DNA length. Another would be that the probability of the two [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] outside ends to synapse to form a transpososome simply decreases with increasing distance between the two.<br /> |
===Other IS''1''-flanked Compound Transposons=== | ===Other IS''1''-flanked Compound Transposons=== | ||
| Line 283: | Line 283: | ||
===[[IS Families/IS6 family|IS''6'' family]] and Pseudocompound Transposons=== | ===[[IS Families/IS6 family|IS''6'' family]] and Pseudocompound Transposons=== | ||
| − | The term pseudocompound transposons (Galas and Chandler 1989 ISBN: 978-1555810054) was coined to describe transposons which are based on [[IS Families/IS6 family|IS''6'' family members]]. The flanking IS are always in direct repeat because [[IS Families/IS6 family|IS''6'' family members]] transpose via the formation of cointegrates followed by a resolution by recombination (Fig. IS6.8). This is treated in detail in the [[IS Families/IS6 family|chapter concerning the IS''6'' family.]] Members of the [[IS Families/IS6 family|IS''6'' family]], in particular, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS26 IS''26''], play an important role in plasmid rearrangements and fusions and in gene amplification. There are many examples of genes flanked by directly repeated [[IS Families/IS6 family|IS''6'' family]] copies. However, although direct transposition of [[IS Families/IS6 family|IS''6'' members]] generate target repeats of 8bp, intra molecular transpositions which lead to deletions and inversions, do not generate the flanking repeats ([[:File:FigIS6.9.png|Fig. IS6.9]]). To determine whether genes flanked by direct [[IS Families/IS6 family|IS''6'' family]] copies are indeed part of a compound transposon in the absence of direct target repeats, it is necessary to identify the same structures in different genetic environments. Many of the [[IS Families/IS6 family|IS''6'' family]]-based compound transposons listed in [https://tncentral.ncc.unesp.br/ TnCentral] which have been assigned Tn identification numbers have not fulfilled either criterion. | + | The term pseudocompound transposons (Galas and Chandler 1989 ISBN: 978-1555810054) was coined to describe transposons which are based on [[IS Families/IS6 family|IS''6'' family members]]. The flanking IS are always in direct repeat because [[IS Families/IS6 family|IS''6'' family members]] transpose via the formation of cointegrates followed by a resolution by recombination ([[:File:FigIS6.8.png|Fig. IS6.8]]). This is treated in detail in the [[IS Families/IS6 family|chapter concerning the IS''6'' family.]] Members of the [[IS Families/IS6 family|IS''6'' family]], in particular, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS26 IS''26''], play an important role in plasmid rearrangements and fusions and in gene amplification. There are many examples of genes flanked by directly repeated [[IS Families/IS6 family|IS''6'' family]] copies. However, although direct transposition of [[IS Families/IS6 family|IS''6'' members]] generate target repeats of 8bp, intra molecular transpositions which lead to deletions and inversions, do not generate the flanking repeats ([[:File:FigIS6.9.png|Fig. IS6.9]]). To determine whether genes flanked by direct [[IS Families/IS6 family|IS''6'' family]] copies are indeed part of a compound transposon in the absence of direct target repeats, it is necessary to identify the same structures in different genetic environments. Many of the [[IS Families/IS6 family|IS''6'' family]]-based compound transposons listed in [https://tncentral.ncc.unesp.br/ TnCentral] which have been assigned Tn identification numbers have not fulfilled either criterion. |
===Integrative and Conjugative Elements (ICE) Driven by DDE Transposases.=== | ===Integrative and Conjugative Elements (ICE) Driven by DDE Transposases.=== | ||
| Line 289: | Line 289: | ||
====TnGBS1 and TnGBS2==== | ====TnGBS1 and TnGBS2==== | ||
| − | These were initially identified by genome comparisons of ''[https://pt.wikipedia.org/wiki/Streptococcus_agalactiae Streptococcus agalactiae]'' which revealed a composite organization composed of a conserved backbone with many (11–14) large variable regions resembling genomic islands <ref><nowiki><pubmed>16172379</pubmed></nowiki></ref>. A later study of eight [https://pt.wikipedia.org/wiki/Streptococcus_agalactiae ''S.'' ''agalactiae'' genomes] identified 35 of these regions including a number of putative '''ICE'''. Circular forms of five of these were detected <ref><nowiki><pubmed>18708498</pubmed></nowiki></ref>. One chromosomal '''ICE''' (originally called X) was also observed to mobilize chromosomal DNA in a similar way to Hfr conjugative transfer <ref name=":33"><nowiki><pubmed>18832470</pubmed></nowiki></ref>. In one strain (NEM316) there were three copies of one putative '''ICE''', pNEM316-1 <ref name=":33" /> whose junctions with the backbone DNA exhibited identical 25bp imperfect inverted repeats and 9-10 bp flanking direct repeats. In two cases, an “empty” chromosome sequence was identified demonstrating that the DRs were generated from the target DNA as a consequence of insertion. A circular species was also identified in which the IR were abutted and separated by 8bp whose sequence could not be determined, presumably because the circles were derived from a number of inserted elements with identical IRs <ref><nowiki><pubmed>12354221</pubmed></nowiki></ref>. The X (33.5 kbp) and pNEM316-1 (47 kbp) elements were subsequently renamed '''TnGBS2''' and '''TnGBS1''' respectively and were found to contain a DDE transposase related to the eukaryotic Mutator-like transposases (MULT) at their right ends <ref name=":31" />(Fig. DDE ICE.1) Further studies showed that this was very closely related to the prokaryotic [[IS Families/IS256 family#ISLre2|IS''Lre2'']]. They were also shown to have flanking IRs. | + | These were initially identified by genome comparisons of ''[https://pt.wikipedia.org/wiki/Streptococcus_agalactiae Streptococcus agalactiae]'' which revealed a composite organization composed of a conserved backbone with many (11–14) large variable regions resembling genomic islands <ref><nowiki><pubmed>16172379</pubmed></nowiki></ref>. A later study of eight [https://pt.wikipedia.org/wiki/Streptococcus_agalactiae ''S.'' ''agalactiae'' genomes] identified 35 of these regions including a number of putative '''ICE'''. Circular forms of five of these were detected <ref><nowiki><pubmed>18708498</pubmed></nowiki></ref>. One chromosomal '''ICE''' (originally called X) was also observed to mobilize chromosomal DNA in a similar way to Hfr conjugative transfer <ref name=":33"><nowiki><pubmed>18832470</pubmed></nowiki></ref>. In one strain (NEM316) there were three copies of one putative '''ICE''', pNEM316-1 <ref name=":33" /> whose junctions with the backbone DNA exhibited identical 25bp imperfect inverted repeats and 9-10 bp flanking direct repeats. In two cases, an “empty” chromosome sequence was identified demonstrating that the DRs were generated from the target DNA as a consequence of insertion. A circular species was also identified in which the IR were abutted and separated by 8bp whose sequence could not be determined, presumably because the circles were derived from a number of inserted elements with identical IRs <ref><nowiki><pubmed>12354221</pubmed></nowiki></ref>. The X (33.5 kbp) and pNEM316-1 (47 kbp) elements were subsequently renamed '''TnGBS2''' and '''TnGBS1''' respectively and were found to contain a DDE transposase related to the eukaryotic Mutator-like transposases (MULT) at their right ends <ref name=":31" />([[:File:FigICE-1.png|Fig. DDE ICE.1]]) Further studies showed that this was very closely related to the prokaryotic [[IS Families/IS256 family#ISLre2|IS''Lre2'']]. They were also shown to have flanking IRs. |
| − | To explore the relationship between '''ICE''' and the MULT group, genes were retrieved by cascade [https://www.ncbi.nlm.nih.gov/books/NBK2590/ PSI-Blast] using as initial query the transposase of '''TnGBS2''' (see [[IS Families/IS256 family#The IS256 cluster|IS256_family/The_IS256_cluster]] <ref name=":32" />. These fell into 5 groups, MULT 1-5, based on their transposase, sequences ([[:File:Fig. IS256.1.png|Fig. IS256.1]]), their active sites ([[:File:Fig. IS256.2.png|Fig. IS256.2]]) and on their left and right ends. Maps of a sample of DDE ICE together with there is relatives are shown in Fig. DDE ICE.2. | + | To explore the relationship between '''ICE''' and the MULT group, genes were retrieved by cascade [https://www.ncbi.nlm.nih.gov/books/NBK2590/ PSI-Blast] using as initial query the transposase of '''TnGBS2''' (see [[IS Families/IS256 family#The IS256 cluster|IS256_family/The_IS256_cluster]] <ref name=":32" />. These fell into 5 groups, MULT 1-5, based on their transposase, sequences ([[:File:Fig. IS256.1.png|Fig. IS256.1]]), their active sites ([[:File:Fig. IS256.2.png|Fig. IS256.2]]) and on their left and right ends. Maps of a sample of DDE ICE together with there is relatives are shown in [[:File:FigICE-2.png|Fig. DDE ICE.2]]. |
[[File:FigICE-1.png|center|thumb|700x700px|'''Fig. DDE ICE.1 Maps of TnGBS1 and TnGBS2.''' The open reading frames are indicated by horizontal boxes. The direction of expression is indicated by the arrowheads. DDE transposase, red; ''virBD'', green; ''rep/parA'', orange; the vertical orange arrow indicates the origin of transfer. The bottom panel shows an IS which is closely related to the DDE transposases.]] | [[File:FigICE-1.png|center|thumb|700x700px|'''Fig. DDE ICE.1 Maps of TnGBS1 and TnGBS2.''' The open reading frames are indicated by horizontal boxes. The direction of expression is indicated by the arrowheads. DDE transposase, red; ''virBD'', green; ''rep/parA'', orange; the vertical orange arrow indicates the origin of transfer. The bottom panel shows an IS which is closely related to the DDE transposases.]] | ||
| − | [[File:FigICE-2.png|center|thumb|700x700px|'''Fig. DDE ICE. 2 ICE with DDE Transposases and their IS Relatives.''' Symbols as for Fig. DDE ICE.1. The five TEs families: '''(A)''' p-MULT 1, [[IS Families/IS256 family|IS''256'' famil]]<nowiki/>y; '''(B)''' p-MULT2,[[IS Families/IS256 family#ISH6|IS''H6'' family]]; '''(C)'''p-MULT3,Tn''GBS''/[[IS Families/IS256 family#ISLre2|IS''Lre2'' family]]; '''(D)''' p-MULT 4, [[IS Families/IS256 family#ISAzba1|IS''Azba1'']], [[IS Families/IS256 family#ISMich2|IS''Mich2'']], and [[IS Families/IS256 family#ISKra4 2|IS''Kra4'']] families; '''(E)''' p-MULT 5 ''[https://pt.wikipedia.org/wiki/Mycoplasma Mycoplasma]'' ICE family. Arrows represent genes.]] | + | [[File:FigICE-2.png|center|thumb|700x700px|'''Fig. DDE ICE. 2 ICE with DDE Transposases and their IS Relatives.''' Symbols as for [[:File:FigICE-1.png|Fig. DDE ICE.1]]. The five TEs families: '''(A)''' p-MULT 1, [[IS Families/IS256 family|IS''256'' famil]]<nowiki/>y; '''(B)''' p-MULT2,[[IS Families/IS256 family#ISH6|IS''H6'' family]]; '''(C)'''p-MULT3,Tn''GBS''/[[IS Families/IS256 family#ISLre2|IS''Lre2'' family]]; '''(D)''' p-MULT 4, [[IS Families/IS256 family#ISAzba1|IS''Azba1'']], [[IS Families/IS256 family#ISMich2|IS''Mich2'']], and [[IS Families/IS256 family#ISKra4 2|IS''Kra4'']] families; '''(E)''' p-MULT 5 ''[https://pt.wikipedia.org/wiki/Mycoplasma Mycoplasma]'' ICE family. Arrows represent genes.]] |
==Bibliography== | ==Bibliography== | ||
Revision as of 19:00, 26 September 2024
Contents
- 1 History
- 2 Properties of Compound Transposons
- 3 Tn10 (see also IS4 family)
- 3.1 Origin
- 3.2 Polar Mutations and the Presence of an Outward-Directed Promoter Tn10.
- 3.3 Adjacent Deletions and Inversions
- 3.4 Deletions, Inversions and Nearly Precise Excision
- 3.5 “Precise” Excision.
- 3.6 Transposition or Replication?
- 3.7 Tn10 Excision and Host Functions
- 3.8 Other Properties of Tn10: Active IE ends, Inverse Transposition
- 3.9 Other Properties of Tn10: The Flanking IS10 are Functionally Different
- 3.10 Other Properties of Tn10: Tn10 Transposition frequency as a function of length (Transposition from a λ prophage)
- 3.11 Other Properties of Tn10: Lack of Length Dependence Using a Plasmid Donor
- 4 Tn5 (see also IS4 Family)
- 4.1 Origin
- 4.2 Precise Excision and plasmid F
- 4.3 Increased Recombination Between Directly Repeated IS.
- 4.4 The fer Mutations are in Previously Identified tra Genes
- 4.5 In trans Tn5 excision, Homologous Recombination and the Involvement of Conjugation
- 4.6 The Inverted Repeats of Tn5 are Functionally Different.
- 4.7 Both flanking IS50 copies are independently Transposable.
- 4.8 An “escape” Promoter in the IS50 Ends?
- 5 Tn903 (see also IS5 Family)
- 6 Tn9 (see also IS1 family)
- 7 Other IS1-flanked Compound Transposons
- 8 IS256 family Transposons
- 9 IS6 family and Pseudocompound Transposons
- 10 Integrative and Conjugative Elements (ICE) Driven by DDE Transposases.
- 11 Bibliography
History
Composite or compound transposons (e.g. [1]) are DNA segments in which copies of the same or closely related IS flank any gene and act in concert in moving the intervening marker. These were identified early in the study of transposition. While there are many potential compound transposons listed in TnCentral, very few of these have been demonstrated to transpose (i.e. found in different genetic environments or with flanking direct target repeats). Below we describe the structures and properties of the more well characterized examples.
Perhaps the earliest observation of transposition of a compound transposon was the acquisition of a chloramphenicol resistance gene by bacteriophage P1 from a multiple antibiotic resistance plasmid, Rms 14, first described in 1964 [2] and thought to reflect the genetic transducing properties of the phage (see also [3]). It was observed that induction of a λ prophage in a strain carrying a P1cam generates λ cam phage in a process independent of the host rec and phage red systems [4]. Furthermore, electron microscope heteroduplex studies showed that a Cam DNA segment of identical length was located at different positions in different λ isolates. This DNA segment was found to be 2.6 kilobases (kb) long and carry the chloramphenicol resistance gene, Cm, bounded by two directly repeated IS1 copies (MacHattie and Jackowski, 1977). This was called Tn9. It was observed that, on further growth, a small proportion of the λ cam phage lose their resistance to Cm but retain a small Tn9 segment representing a single IS1 copy, presumably the result of recombination between the two directly repeated flanking IS1 copies (Rosner and Gottesman 1977) (Fig. Comp.1).
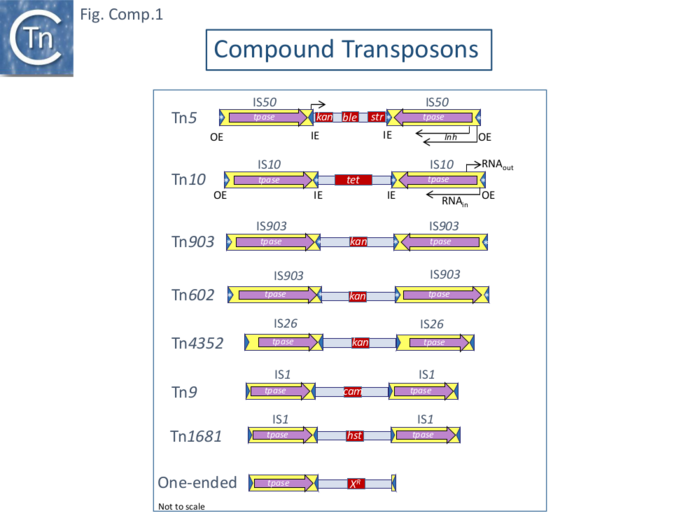
Although Tn9 is bordered by directly repeated IS copies, other transposons were identified at about the same time in which the resistance gene was flanked by inverted copies of an insertion sequence: they included Tn5 carrying a kanamycin resistance gene flanked by copies of the IS50 insertion sequence which generates characteristic “snapbacks” that were detected by microscopy following denaturation and renaturation of vector DNA carrying the insertion (Fig. 2.4) [5], and Tn10 [6][7] whose flanking IS were originally thought to be IS3 but which were later shown to be copies of IS10 [8] and which carries resistance to tetracycline (Fig. Comp.1).
Other early examples are: Tn1681, composed of a heat stable toxin gene flanked by directly repeated copies of IS1 [9]; Tn903 (Fig. Comp.1) (also known as Tn601 and Tn55 [5] [10]; and Tn603 [11].
These early examples indicated that: 1) the flanking IS can be found direct or inverted with respect to each other (e.g. Tn9 and Tn10); 2) the same interstitial gene can be flanked by identical IS in direct or inverted orientation e.g. IS602 and IS903; 3) the same gene can be flanked by different IS in different transposons (e.g. APH(3’)-Ia : Tn903 flanking IS903 and Tn4352 with flanking IS26 [12]; and 4) the same IS can flank different interstitial genes (e.g. Tn9 - Cmr and Tn1681 - heat stable toxin).
Properties of Compound Transposons
Compound Transposons Exhibit Additional Properties to Flanking Insertion Sequences Alone
Most analyses of transposition have, understandably, focused on the IS themselves since many of these are found individually as components of chromosomes and plasmids (see [13]) and can transpose independently. However, compound transposons themselves exhibit properties that the individual IS do not. The behavior of the complete transposon has been investigated in only a limited number of cases, notably Tn5, Tn9, Tn10 (Fig. Comp.1) and Tn903.
Tn10 (see also IS4 family)
Origin
The tetracycline, tet, resistance genes from the plasmid R100 (also known as NR1 and R222)(Fig. IS1.2) and a number of other R plasmids were capable of transposition from one replicon to another in the absence of the host recA homologous recombination system [7][14] . Insertions of the transposon, Tn10 [15], were found to be mutagenic when occurring in structural genes and polar when inserted into operons [7][16]. Tn10 insertion generates a short 9bp target repeat flanking each transposon end [17] and shows a preference for the sequence GCTNAGC located symmetrically within the 9bp repeat [18].
Partial Tn10 restriction maps had been published but the Reznikoff lab generated a complete physical restriction map in 1979 [19] and partial sequences of the flanking IS10 elements were initially published [20] but it was not until 2000 that a full Tn10 sequence became publicly available [21] (Fig. Tn10.1).
Although transposition and its regulation has been focused on the individual flanking IS10 elements (see [22]) and is described in the IS4 family chapter, Tn10 exhibits a number of properties which the flanking IS on their own do not. Much attention has been focused on Tn10-promoted genome instabilities stemming from its capacity generate adjacent deletions and to undergo excision. Understanding these for Tn10 as well as other transposons is key to understanding prokaryotic genome evolution.
Polar Mutations and the Presence of an Outward-Directed Promoter Tn10.
A Tn10 copy acquired by phage P22 was observed to insert into many places in the Salmonella typhimurium chromosome generating a variety of auxotrophs. Insertion into an operon was found to be polar on downstream genes and a number of insertions were identified in the Salmonella his operon [7]. The complementation studies were consistent with the presence of two additional internal promoters described by Atkins and Loper 1970 [23] with the organization:
P1-hisO-hisG-hisD-hisC-P2-hisB-hisH-hisA-hisF-P3-hisI-hisE.
Insertions into the P1 promoter-proximal hisG prevented expression of the adjacent hisD and hisC genes while genes located downstream to hisC are expressed from a second promoter, P2, between the hisC and hisB genes and insertions in hisB downstream of P2 prevent expression of the hisH, hisA, and hisF genes. The hisI and hisE genes are transcribed from the secondary promoter P3 located at the distal end of the hisF gene.
Tet sensitive revertants were identified but only a minority had regained their his prototrophy. Another study analyzed the Tn10 copy from R100 into the E. coli lac operon and also demonstrated that insertion into lacZ was polar on lacY, that there was a preferential site or region for insertion at the 3’ end of the gene lacZ gene and that reversion to lac+ occurred at low frequencies [24].
Although most his complementation data fitted the expected pattern, the hisH::Tn10 mutants showed weak expression of the downstream hisA and hisF [7]. Further analysis [25] showed that this was due to a promoter within Tn10 and that, in the absence of the transcription terminator, ρ, the downstream genes were expressed. This implies that the transcript escaping from Tn10 is not translated. Otherwise this would prevent ρ activity. The Tn10 promoter responsible is presumably Pout (Fig. IS4.3) involved in production of antisense RNA (RNA-OUT) involved in regulation of IS10 transposase expression (see also: Small RNAs from IS4 family chapter)
Adjacent Deletions and Inversions
Early studies also showed that tetracycline sensitive mutants of R100.1 whose tra operon is adjacent to Tn10 on the plasmid were often deficient in conjugative transfer [26]. Tn10 excision (tetS), when inserted into various locations in the transfer (tra) operon of R100, also generated adjacent deletions and probable inversions (Fig. Tn10.2) [27][28].
Kleckner [17][29] demonstrated that insertion of Tn10 generated flanking 9bp direct target repeats on insertion and that these repeats were not necessary for further transposition. Moreover, in addition to adjacent deletions, Tn10 excision was found to promote inversion events [30] when inserted into the his operon in the S. typhimurium chromosome.
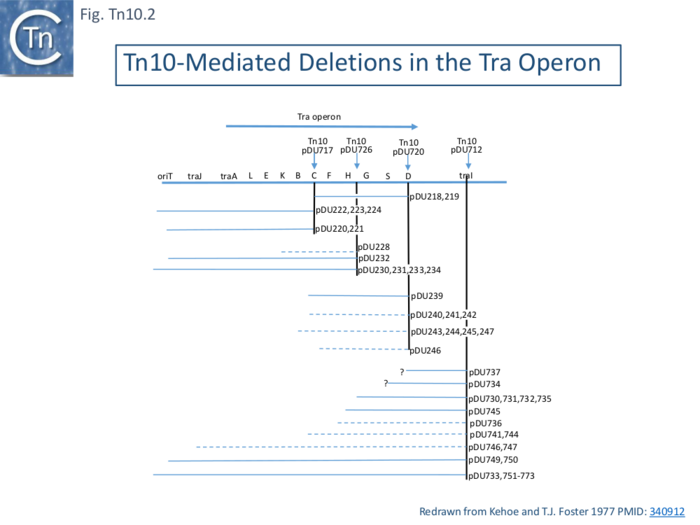
Deletions, Inversions and Nearly Precise Excision
Deletions in a bacteriophage λ with a Tn10 insertion, λ::Tn10, following a round of induction and deletion enrichment using resistance to EDTA treatment (phage with an increased genome size are sensitive to EDTA) followed by replating to obtain tetracycline sensitive phage, identified three classes as judged by restriction site and heteroduplex mapping: 1) simple deletions; 2) deletion accompanied by an inversion; and 3) nearly precise excisions retaining a small 50bp segment of the outside ends of the flanking IS10.
Deletion was independent of the phage and host homologous recombination systems (rex and rec) The structure of these deletions was determined by a combination of electron microscopy heteroduplex analysis and restriction mapping.
Five of 26 deletions (#250, 260, 261 264 and 317; Fig. Tn10.3 A) appeared to have retained the flanking IS10 and were missing parts of the interior tet region. All had deletion endpoints which mapped to one or other or, in the case of #317, both of the IS10 IE. Three were found to have deleted one or other of the flanking IS either partially (#267) or fully (#270, 265).
The second class of deletion was accompanied by small inversions. Six deletion derivatives were analysed in detail ( Fig. Tn10.3 B). In all, the intervening DNA between the flanking IS was deleted. These types of deletion and inversion/deletion have been proposed [28] to occur by intramolecular transposition using both flanking IS10IE (Fig. Comp.1) deletions occur when insertion occurs into the target in one orientation (Fig. Comp.5 left) while inversion/deletions occur when insertion occurs in the opposite orientation (Fig. Comp.5 left). The inversion/deletion events should give rise to circular products of the deleted DNA but, since they are unable to replicate, are lost during cell growth. Such circular products had subsequently been observed [32]. They proved to be interrupted in one or other of the DNA strands and were interpreted as abortive transposition events in which the transposon had inserted within itself producing abutted left and right ends. A later study [33] demonstrated that these were in fact deletion/inversion products in which the transposon ends had inserted in targets within the transposon.
The third type of tetS deletion, analyzed in an associated study was called “nearly precise excision” ( Fig. Tn10.3 C ) [34]. These appeared to have occurred at AT-rich sub-terminal inverted repeats located at the outside end (OE; Fig. Tn10.3 C) [20] which are involved in the RNAin/RNAout antisense RNA regulation of transposase expression [35][36][37].
Tn10-mediated Deletions and Deletion/Inversion are proposed to be generated by intramolecular transposition using Tn10 IE ( Fig. Tn10.4) which insert into the target DNA in one orientation to generate deletions (Fig. Tn10.4 left) and in the other to generate inversions/deletions (Fig. Tn10.4 right). In both cases, this leads to loss of the tetR gene.
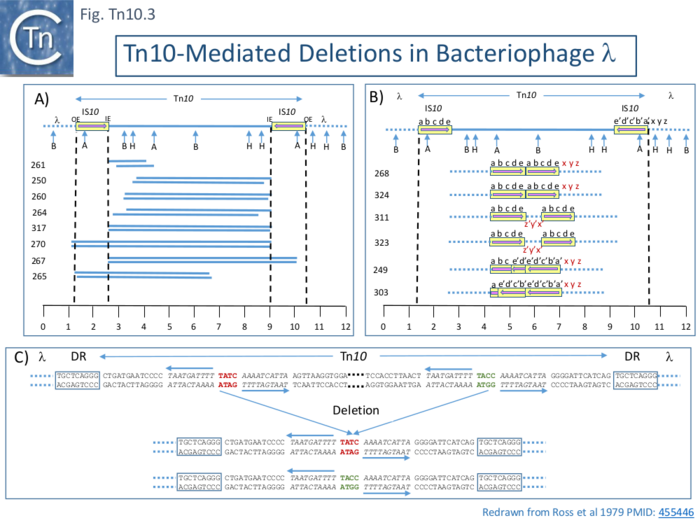
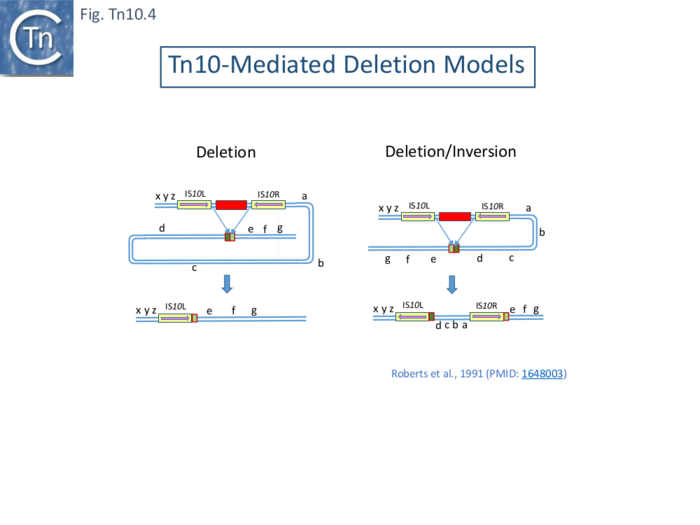
It should be noted that using phage λ as a deletion test system is accompanied by certain constraints: since isolation of the deletants occurred after phage replication, it is possible that this type of deletion involves the passage of a replication fork; and long deletions such as those identified in the plasmid F-tra operon (Fig. Tn10.2) were not found, presumably due to constraints on phage viability associated with phage size and/or the presence of neighboring essential phage genes.
However, in all three types of deletion, it appeared that the majority occurred using the internal ends of the flanking IS (IE; Fig. Tn10.3), suggesting that these ends, like the OE, are also active.
“Precise” Excision.
Foster et al [39] identified a number of Tn10-mediated events in addition to the adjacent deletions and nearly-precise excision (Fig. Tn10.3). These, which all generate new joints between the transposon end and a target sequence, are precise excision of the entire Tn10 and precise excision of the Tn10 remnant remaining after nearly precise excision. The novel deletions and nearly-precise excision all involved small direct repeats at or near the Tn10 ends resulting in deletion of all intervening DNA but leaving one of the repeats in place. The DNA sequence of a his+ revertant of a his::Tn10 derivative of S.typhimurium revealed a wildtype sequence with a single copy of the direct target repeat generated by the original Tn10 insertion event. The his system also generated nearly precise excision events using subterminal repeats in OE similar to those found in the lambda system (Fig.Tn10.3 C). These physical structures are consistent with previous genetic results in which a Tn10 insertion into hisG (hisG::Tn10) is polar on the downstream hisD [30]. Two types of hisD polar-relief revertants were obtained. In one class, the authors could find little or no Tn10 homologous material (with a resolution of >50 bp by electron microscopy) but these could subsequently give rise to full his+ offspring. This can now be interpreted as a twostep process with an initial nearly precise excision, which leaves 50 bp of Tn10 (Fig.Tn10.3 C) and is non-polar on hisD, followed by a precise excision of the remaining Tn10 material to generate a his+ phenotype.[1]
Moreover: Tn10 derivatives defective for transposition are unaffected in their excision and nearly precise excision frequencies; excision and nearly precise excision frequencies decrease with Tn10 deletion derivatives carrying shorter inverted repeats (IS10) [39]. Similar conclusions were reached for Tn5, which is structurally similar to Tn10 (Fig. Comp.1), strongly suggesting that long inverted flanking IS are key (for Tn5 see also Egner and Berg [40]).
Rearrangements promoted by a single chromosomal insertion of Tn10 or an artificial TnGal in E. coli and S. typhimurium were analyzed by Southern blot hybridization of DNA isolated from many individual clones [41] and digested with the appropriate restriction enzymes. This provided physical evidence of transposition events by following the retention or loss of Tn-chromosome junctions and IS10-internal transposon junctions. The authors were able to estimate transposition frequencies of the entire transposon and of the individual flanking IS: transposition of an individual IS10 was 10-4 per cell per generation in overnight cultures grown and plated on minimal media, a value at least ten fold higher than other Tn10/IS10 promoted events (e.g. deletions etc); all chromosomal rearrangements were either deletion/inversions or deletions as had been previously analyzed for Tn10; the rearrangements were at least 10 fold lower than IS10 transposition; and Tn10 events used nearby target sites while IS10 events appeared to target “randomly” around the chromosome.
Transposition or Replication?
It was suggested that these deletions may be generated during the transposition process when two IS10OE ends (IS10L and IS10R) are synapsed which would explain why excision is less frequent than transposition. An alternative explanation would be that excision occurs during replication when a replication fork passes through the transposon. This might allow pairing of IS10L and IS10R to form “snapbacks” (see Fig. 2.4) on the discontinuous (lagging) strand, facilitating replication slippage between the two directly-repeated sequences. This explanation has been proposed in the case of precise excision of Tn5.
Tn10 Excision and Host Functions
A number of host mutants (~40) in which precise and nearly precise excision was increased were isolated following nitrosoguanidine (NG) mutagenesis of E. coli carrying a lacZ: :Tn10 chromosomal insertion and a readout of increased lac+ papillae in single colonies [39][42][43].
While affecting precise excision, these mutations, called tex for Tn10 excision, did not appear to affect the precise excision of the Tn10 remnant remaining after nearly precise excision. An initial group were called texA-E and proved to be alleles of recB and/or recC, uvrD, mutH, mutS, and dam, genes involved in host DNA metabolism. It was suggested that precise excision of the nearly-precise excision remnant occurs by a different pathway.
Further genetic mapping of the tex mutants showed that three texA mutants which shared two phenotypic properties in addition to increased precise excision, inhibition of Red- Gam - λ phage growth and sensitivity to UV irradiation characteristic of recBC mutants, were also genetically linked to recBC: texA343 near recC and texA344 near recB and texA345 linked to thyA [44]. These mutations also stimulated precise excision of Tn5.
A further 21 tex mutants exhibited a mutator phenotype and mapped to the uvrD, mutH, mutL, mutS and dam genes, part of the methylation-directed base pair mismatch repair pathway. In addition, existing alleles of these genes also exhibited the Tex phenotype [43]. However, mutT, polA, ung which affect the spontaneous mutation frequency, uvrA, uvrB involved in other excision repair pathways, and dcm which affects the state of cytosine DNA methylation had no effect.
Subsequent studies have implied the involvement of a large number of genes associated with DNA metabolism and processing as well as treatments which lead to higher levels of DNA repair functions. These include: UV light or mitomycin C which increased precise excision from the Salmonella typhimurium met::Tn10, trp::Tn10 and srl::Tn10 genes and was dependent on recA [45].
Low doses of UV light also increased precise excision of both Tn10 and Tn5 from the chromosomal insertion sites ade128 and cyc750 but not the structurally different Tn3 family member, Tn1 in Escherichia coli K-12 and E. coli B cells, wild-type for DNA-repair [46]. However, mutations of the lexA, recA and umuDC genes involved in UV-induced SOS-mutability were reported to eliminate precise excision activity.
Other studies using Tn10 insertions in the E. coli leu::Tn10, thr::Tn10 or gal::Tn10 genes revealed that the lexA3 (ind-) mutation (which prevents auto-cleavage of the SOS network repressor, lexA, and prevents SOS induction [47] eliminated excision in either a recA+ or a recA730 (a constitutively active recA derivative) genetic background, indicating that expression of another SOS component other than recA is required for UV excision induction [48]. This study also indicated that a umuC mutant (Tn5-inactivated gene, umuC::Tn5) had no effect.
An increase in excision frequency of about 20 or 150 times Salmonella typhimurium polA mutants was also detected from srl202:: Tn10 [49], with smaller increases in mutH (2 fold) and uvrD (15 fold) mutants (this study also found increases in transposition in with polA, recA and uvrD mutants using mating out assays). Various components of the host rec system have also been implicated (recG and ruv: [50];. recN: [51]; recA and recF: [52]; recBC and recF: [53]).
Essential Take-home Messages Concerning Tn10 Excision
The important points to note from this ensemble of studies is that: 1) precise and nearly precise excisions occur at short directly repeated sequences; 2) does not depend on the transposition activity of the flanking IS10 sequences; 3) depends on the length of the inverted IS10 sequences; 4) mutants which are selected for increased excision map in genes associated with DNA processing; 5) pre-existing mutants in a large number of DNA processing genes also affect excision.
It is unclear whether these genes intervene prior to the excision event or post-excision to repair. Unfortunately, biochemical analyses are not at present available to provide more detailed information.
Other Properties of Tn10: Active IE ends, Inverse Transposition
That the inside ends are active in transposition as implied in the deletion data (Fig. Tn10.3) was also demonstrated by the observation that integrative suppression (the rescue of chromosome replication in a temperature sensitive dnaAts initiation mutant by plasmid integration) using the R100.1 plasmid (a mutant derivative of R100 derepressed for conjugal transfer) to integrate and drive chromosome replication [54] resulted in the frequent appearance of tet sensitive temperature resistant clones (Chandler et al., 1978 CSH Symp. Quant. Biol. 43). Further analysis demonstrated that in these cells, R100.1 had integrated into the host chromosome by transposition using the IE of both IS10 with the loss of tetR. This was termed “inverse” transposition [55]. Similar use of IE was subsequently reported in the case of a circular plasmid carrying and ampicillin resistance gene together with a Tn10 which gave rise to two types of symmetrical transposition event: normal Tn10 transposition of tetR using both OE and another involving ampR and both opposite, IE, ends in which tet was lost [56].
Other Properties of Tn10: The Flanking IS10 are Functionally Different
Substitution of the internal 6500 bp segment of Tn10 with an alternative resistance gene and deletion analysis of Tn10 (Foster et al 1981 PMID: 6260375) was used to demonstrate that the flanking IS contained all the information necessary for Tn10 transposition. It also revealed that the flanking IS were not functionally equivalent: in derivatives with one or other flanking IS disabled by small insertions or deletions across Tn10, the right hand IS10 (IS10R) was found to be fully functional and to efficiently complement the left hand IS10L while IS10L functions very poorly to complement an inactivated IS10R (a factor of 25-100-fold lower efficiency). Furthermore, all sites necessary (in cis) for transposition were located to within the terminal 70bp of the component IS10. These studies also revealed that unlike the deletion mutants, point mutants defective in transposition could not be complemented in trans. This raises the question of whether the transposase acts preferentially in cis (refs) in which an inactive transposase produced from the point mutants produced in cis inhibits the activity of the wildtype transposase acting in trans.
The difference in activities of IS10L and IS10R were subsequently revealed from sequence data (Halling et al., 1982 PMID: 6283536) where a number of small sequence differences were observed. Those in the coding region were either synonymous or changes to related amino acids (e.g. single examples of I to V, S to L, T to A, or V to A). Sic of the 15 changes were also identified in the upstream regulatory region which includes features such as the promoters pIN and pOUT (IS4 family/IS10) and two sets of inverted repeats.
Other Properties of Tn10: Tn10 Transposition frequency as a function of length (Transposition from a λ prophage)
The frequency of Tn10 transposition was observed to decrease exponentially with increasing length (distance between the two flanking IS10) (Morisato et al 1983 PMID: 6299577) as had previously been observed for Tn9 (Chandler et al. 1982 PMID: 6281439). This was measured with a mating out assay using an F’lacpro plasmid as a target and a series of isogenic Tn10 elements carried in a phage hisG::Tn10 λ prophage whose length was varied largely by deletion of parts of the internal DNA segment to generate derivative of between full-length (9147bp) and ~4500bp. It was stated that similar result was obtained using a hop assay. In a first set of experiments, Tn10 derivatives with active transposase were used. In this case, transposition was said to be “self-driven” (Fig. Tn10.5 A). Transposition frequencies decreased by a factor of 40% for every 1kb increase in transposon length. Moreover, a curve with a similar slope but with higher absolute frequency values was observed if Tn10 transposase was supplied in trans from two different multicopy plasmids to Tn10 derivatives with non-functional transposase (Fig. Tn10.5 A). Plasmid pNK290 contains an IS10R with a regulatory mutation which increases Tn10 transpositon 100 fold while in pNK432, the transposase gene is driven by the histidine operon promoter. The dotted arrows show derivatives which are more active or less active than expected for unknown reasons.
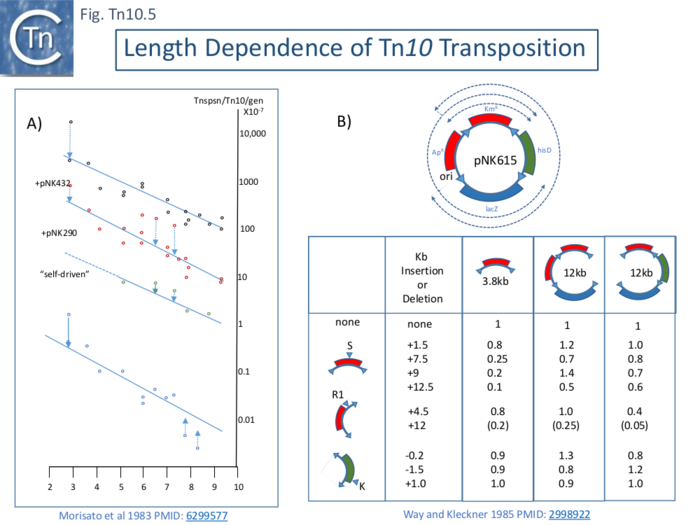
As suggested for Tn9 derivatives, the reduction in activity associated with increasing length may reflect a reduction in probability that the flanking ends can synapse as might be needed for transposition. This might be either the result of random collision of the IR or their meeting via a directional tracking mechanism.
Other Properties of Tn10: Lack of Length Dependence Using a Plasmid Donor
A second set of experiments [58] indicated that length dependent transposition does not occur if circular plasmids are used as donors. (WAS THIS PREDICTED BY US???). Using a plasmid, pNK615, which carries a number of appropriately oriented IS10 ends (Fig. Tn10.5 B Top) as a donor in a mating out assay. The frequencies of transposition of each segment cannot be directly compared since factors such as impinging transcription and DAM methylation in the IR can profoundly influence transposition. To overcome these limitation, various insertions or deletions were introduced into one of three positions creating large variations in length (Fig. Tn10.5 B Bottom) and transposition frequencies were normalized to the non-modified pNK615 segments. Only small differences were noted.
This is perhaps not unexpected since presumably, the transposition frequency would be determined by the shorted distance between the appropriately oriented ends rather that solely the distance in the transposon (see Chandler et al., [59] for discussion)
Tn5 (see also IS4 Family)
Origin
Tn5 (Fig. Tn5.1) was isolated by transposition to bacteriophage λ from the Klebsiella resistance plasmid JR67 which had been transferred to E. coli [5]. It is structurally similar to Tn10 with two long inverted flanking repeats of about 1.45kb. These were subsequently called IS50. Like Tn10, it transposes at relatively high frequencies, at many different sites and is mutagenic, generating polar mutations in operons on insertion.
Precise Excision and plasmid F
Hopkins et al [60] observed that the presence of an F’lacpro (F’128, an F’ generated by excision via two rep sequences; Fig. IS200.72; [61]) plasmid led to a moderate increase in the frequency of reversion of auxotrophic mutants caused by the insertion of Tn5 (and Tn10). The stimulation of excision from Tn5 insertions from chromosomal ilv::Tn5 and pyr::Tn5 was observed to be modest, between 4 and 20 fold. Excision of Tn5 from a lacZ::Tn5 located on a plasmid compared to the same insertion located in the chromosome was even higher: 153 fold. A similar high Tn5 excision frequency was substantially higher from an F’lacZ::Tn5 plasmid than from the chromosome (where they were unable to observe significant excision)(Egner and Berg, 1979 cited in [62]. Plasmid F mutants which had lost the ability to stimulate excision after ethyl methanesulfonate mutagenesis were isolated using a lac papillae screen on Maconkey lac plates[60]. Two low-papillation mutants no longer stimulated precise excision of chromosomal Tn5 insertions. These were called ferA for F factor-mediated excision and recombination. Another two were identified after Tn10 insertional mutagenesis. A second class of mutant which stimulated precise excision, ferB, was found following UV or transposition mutagenesis. Additionally, 3 mutants that stimulated precise excision of Tn10 from lacZ::Tn10 were isolated after insertion mutagenesis with Tn5. The ferB mutants stimulated excision by a factor of 3-6 fold compared to the unmutagenized plasmid. It was proposed that ferB mutants allow overproduction of an excision promoting ferA product and, indeed ferA/ferB double mutants exhibited the ferA phenotype. It was proposed that the ferB mutants occurred in a structural gene rather than in a promoter based on the frequency of UV mutagenesis and the fact that they could also be obtained by insertional mutagenesis.
Increased Recombination Between Directly Repeated IS.
Surprisingly, the F’lacpro plasmid appeared unstable with the ferB mutation – it lost its lac-pro genes but remained F+ - and stability was restored in a recA strain. This was found to be due to recombination between two directly repeated copies of the insertion sequence IS3 which flank the genes. This behavior is reminiscent of the acquisition of the related R100.1 plasmid to produce circular copies of its resistance determinants (r-det) by recA dependent recombination between two flanking directly repeated IS1 (r-det). The two genes were located on the genetic map of the F plasmid.
The fer Mutations are in Previously Identified tra Genes
Subsequent studies [63] showed the fer mutations to be in the tra operon in previously identified tra genes. Using cloning and complementation, with an F'lacZ::Tn5 pro plasmid, ferB was found to be equivalent to traS. TraS is an inner-membrane protein involved in surface exclusion [64], a phenotype that prevents F+ cells from acting as recipients in F mediated conjugation. TraS mutants therefore enhance transfer. The traS stimulatory effect on precise excision should be eliminated in transfer deficient F mutants. Three independent Tn10 insertion mutants, all in different regions of the tra operon, generated the ferA phenotype as did previously characterized mutants in traJ, traI and traD when introduced into the F’. Using genetically distinguishable donor and recipient cells, it was shown that precise excision occurred principally in the recipient following conjugal transfer. It is noteworthy that F’128 and the F’128traS mutant stimulate excision to roughly the same extent in the recipient – thus reinforcing the notion that the increase in excision observed in the F’128traS mutant results from its increased conjugative activity rather than a direct effect of the TraS protein.
In trans Tn5 excision, Homologous Recombination and the Involvement of Conjugation
The stimulation observed between directly repeated IS3 copies [60] was investigated further using a dimeric plasmid target which is unable to be mobilized and cannot be transferred between cells. The plasmid carries different mutations in each of its two tet genes. Recombination between the two genes gives rise to tet resistance and this was found to be 6 fold higher in a strain carrying F'128 traS12::Tn5 compared to a strain with F' in a process which depends on recA. Interestingly, stimulation is even higher in a recF strain suggesting that recombination is dependent on recA but that the recF pathway may compete in some way with the recA pathway. Stimulation of Tn5 excision from chromosomal sites is also an in trans activity.
This study also showed that not all plasmid F derivatives could stimulate excision of chromosomally inserted Tn5 copies in trans. This was true even for a traS mutant F plasmid derived from F’128traS. It appears as though only F’ plasmids carrying large chromosome segments exhibit in trans excision stimulation suggesting that Hfr formation and conjugation may be occurring in these cases. In some or the original papers concerning the fertility factor, F, it was observed that various E.coli chromosomal genes were each transferred independently at very low frequencies (see [65]) this was found to be the result of F integration into the chromosome and subsequent transfer of neighboring chromosomal genes. F’ can integrate into the chromosome at higher frequencies due to their significant similarity. It is therefore presumably this effect and the subsequent conjugation which explains the “trans” effect of various F’ to stimulate chromosomal Tn5 excision.
Similar conclusions were drawn by the Berg lab [62] who showed that Tn5 was excised from an F’lac::Tn5 plasmid several orders of magnitude more efficiently than from its chromosomal location in F-, F+ or even from a homozygous strain carrying the F’lac::Tn5. In these experiments, significant transfer was also observed into a strain carrying an F’. reinforcing the idea that conjugative transfer is necessary for the stimulation.
In conjugative replication, the transferred strand is replicated as the non-continuous strand in the recipient and excision is probably facilitated by the (partially) single strand nature of the transferred DNA allowing “snapback” formation (see Fig. 2.4).
The idea that replication of single stranded DNA leads to Tn5 excision is reinforced by the observation that the single strand phage, fd, can accommodate the insertion of Tn5 but loses it at a high rate on propagation [66].
The Inverted Repeats of Tn5 are Functionally Different.
Like those of Tn10, the flanking Tn5 repeats are functionally different [67]. An extensive restriction endonuclease map (Fig. Comp.7) [19] was used to design a library of Tn5 mutants. This localized the aph gene proximal to the inside end (IE) of IS50L. Insertions at the left BglII site were found to severely reduce neomycin resistance levels [19] suggesting that aph expression could be driven from IS50L. This was confirmed by the observation that inversion of a BglII fragment resulted in loss of resistance while resistance is retained on inversion of a HindIII fragment (Fig. Tn5.1) [67].
RNA polymerase binding sites were located using gel mobility shift assays: binding occurred in DNA fragments at both outside ends (OE) and a third weak binding site was located close to IE of IS50L but not in this location in IS50R (Fig. Tn5.1). The direction of transcription was determined in vitro using different size fragments and determining the length of the run-off transcripts. It was found to start at OE and continue through the IS. Surprisingly, two distinct transcripts were identified differing at their 5’ end by about 10 nucleotides.
Five distinct Tn5-specific translation products were identified using a minicell system (of 58K, 54K, 53K, 49K and 26K). A major product had a mass, 26Kd, consistent with that expected for the neomycin phophotransferase. The sum of the other 4 species was clearly significantly larger than the coding capacity of IS50 elements. The smallest of these species, 49Kd, had previously been shown to be encoded by IS50L [68]. An insertion into the HindIII site of IS50L resulted in the loss of the 53Kd and 49Kd species whereas an insertion into the HindIII site in IS50R resulted in the loss of the 58K and 54K species [67]. The assignment of the species to IS50L or IS50R was confirmed by deletion of one or other of the IS. Concomitant with the loss of the 4 protein species, insertions in the HindIII sites of IS50L and IS50R both gave rise to species of 42K and 38K, fusion protein products resulting from the inserted DNA. These results imply that the difference in size of the two proteins from one IS derives from a difference in their N-terminal ends whereas the difference in size of the proteins between the two IS results from their C-terminal ends (Fig. Tn5.1). To further localize the open reading frames, a Tn5 derivative deleted for both ends at the HpaI-HindII site does not produce the 58K and 53K species, since the RNA pol binding site is removed, and produces reduced amounts of the 54K and 49K proteins (probably due to impinging transcription from the surrounding sequence context).
To determine whether these differences between IS50L and IS50R are reflected in their abilities to drive Tn5 transposition, a number of mutations were introduced into either IS50L or IS50R. Ablation of IS50R consistently had a severe effect on transposition of the Tn5 derivative whereas ablation of IS50L had only a small effect. IS50R makes a major contribution to Tn5 transposition while IS50L is necessary for expression of the neomycin resistance gene.
This difference in activity of IS50L and IS50R is the consequence of a single base change in IS50L which creates a better promoter sequence for the expression of neomycin resistance and introduces a UAA nonsense codon which can be suppressed by a suitable ochre suppressor rendering the truncated proteins functional in transposition [69].
Both flanking IS50 copies are independently Transposable.
As a first step in demonstrating that IS50L and IS50R are each active IS, it was shown that IS50 IE (inside ends), like the outside ends (OE) were capable of contributing to transposition. This was achieved by examining the capacity of Tn5 to undergo inverse transposition (using the IS50 IE) as does Tn10. For this, inverse transposition events from a Δ::Tn5 to pBR322 were scored as KmS (loss of the Tn5 gene), ApR,TetR (pBR322) transducing phage. Twenty independent potential Δ::pBR322 cointegrates were analyzed by restriction mapping and shown to carry pBR322 and to have generated new IS50- Δ junction fragments whose size was different in each isolate [70].
Furthermore, transposition of the IS50L, IS50R and Tn5 components were measured individually using derivatives of Tn5 inserted into a chromosomal lac gene in which either the left or right IS50 was marked by insertion of a Tet gene. A 2700 bp TetR DNA fragment was inserted into the BglII site which does not affect the ability of IS50 to transpose but in view of its size might affect transposition frequency (e.g. Tn10 Transposition frequency as a function of length; Tn9 Transposition frequency as a function of length). The transposition target used was phage λ which prevents scoring of inverse transposition from the chromosome since it would not result in viable phage. TetR KmS (kanamicyin) transducing phage was used as a measure of IS50 transposition and TetR KmR as a measure of Tn5 transposition. It was observed that 10% and 30% of TetR phage were KmS using Tn5 transposons with marked IS50L or IS50R respectively which was a measure of transposition of the marked IS50 while the remainder of the TetR phage were also KanR indicating transposition of the entire Tn50. Thus both flanking IS50 can transpose independently.
An “escape” Promoter in the IS50 Ends?
Although Tn5 insertion is largely polar, it was observed that some low level of downstream expression occurred in a portion of insertions into the lac gene [71]. This did not depend on the orientation of Tn5 – which could be inverted without affecting this expression leakage. The authors suggest that it is within the terminal ~180bp. It should be noted that it unlikely to be equivalent to the Pout (Fig. IS4.3) promoter identified in the case of IS10. Retrospectively, it could be due to the formation of hybrid promoters with a -35 promoter component supplied by the transposon and a -10 component present in the neighboring target DNA as has been noted for a number of insertion sequences (see: General Information/IS and Gene Expression).
Tn903 (see also IS5 Family)
Origin
Tn903 was originally isolated from plasmid pML21 [72] (a hybrid plasmid consisting of pVH51 and the KmR fragment of plasmid pSC105 inserted at the EcoRI restriction site of mini-ColE1), by transposition onto the phage fd [73] and shown to transpose to multiple sites in other replicons. Tn903 transposition activity is low, and it has previously been used as a stable selective marker on many plasmids used as cloning vehicles. It is carried by plasmid R6(-5) [74]. It was shown to comprise a central region of about 1kb carrying a kanamycin resistance gene flanked by 1050bp inverted repeats [73] (Fig. Tn903.1) which were called IS903 [75]. Insertion results in a target duplication of 9bp but insertion sites did not appear to have any common sequence features [10]. Deletion of an internal restriction fragment from both repeats eliminated Tn903 transposition but deletions of the fragment from only one repeat retained transposition activity. Moreover, an isolated IS903 copy was shown to undergo independent transposition [75]. The complete nucleotide sequence was obtained by Oka et al [76] who identified the kanamycin resistance (APH) gene and its product in a minicell system.
Direct and Replicative Transposition.
Investigation into Tn903 transposition has not been as extensive as for Tn5 and Tn10. However, one property that distinguishes Tn903 from Tn5 and Tn10 is that it may to be able to undergo replicative transposition as well as direct transposition [77][78][79][80].
The Syvanen lab [81][82] using a phage lambda delivery system observed both direct transposition events into the E. coli chromosome, similar to those observed with Tn5 and Tn10. However, they also observed integration of the entire vector at a specific point in the chromosome. However, unlike Tn5 and Tn10 events, both appeared to require the recA function. Further analysis indicated that insertion into the lambda genome resulted in the production of defective λdv phage containing large rearrangements presumably generated by the Tn903 insertion. It seems possible that this behavior is linked to the particular λ vector used. No further information has become available.
Cointegrate formation?
Replicative transposition, is often evidenced by the formation of cointegrates. However, it should be underlined that identification of cointegrate structures does not necessarily indicate replicative transposition since they can be formed by direct transposition from a dimeric donor plasmid (Fig. Tn903.2 A where ii is a direct transposition and v an inverse transposition while both iii and iv resemble replicative cointegrates but in fact are formed by direct transposition). Plasmid dimerization can occur even in the absence of the recA function [83]. The monomer-dimer plasmid status might also be impacted by the recombination status of the host although plasmid dimerization can occur even in the absence of the recA function [83]. This had been noted in the case of Tn5 [84] where cointegrate formation was found to be dependent on the recA function [85] and was ascribed to direct transposition from plasmid dimers [86].
Replicative Intramolecular Transposition
To address the question of replicative transposition of Tn903, Weinert et al [77][78] investigated intramolecular transposition using a cloned neighboring galK gene copy next to Tn903 in a pBR322-derivative plasmid. High galactokinase (GalK) expression is lethal in an E. coli galE-T- strain in the presence of galactose. Selection of galactose resistance selects for mutation/deletion/rearrangement of galK. Deletions using a highly active IS903 retained the IS and eliminated DNA from through into and through galK as would be expected to result from an intramolecular replicative cointegrate formation with the IS903 repeat joined to a new target sequence (see: Tn3 family and IS6 family). It was further shown that deletion require both IS903 ends. Using a cointegrate formed between the galK pBR322 plasmid and pOX38, it was possible to identify both parts of the deletion product, since each carries a replicative origin, using their different drug resistances (Fig. Tn903.2 B). Additionally, intramolecular replicative transposition should generate a direct target repeat at the insertion point which would abut each new junction in each deletion product. The authors identified such paired 9bp sequences as expected providing strong evidence for a replicative transposition process (Fig. Tn903.2 B).
Mutation and a Balance between Direct and Replicative Transposition
Further insights were provided by Tavakoli and Derbyshire [80] who demonstrated that the nucleotides directly flanking IS903 (Tn903) influence the ability to undergo replicative cointegrate formation. Additionally, they isolated transposase mutants which also showed an increase in cointegrate formation. They suggested that normally the IS903 (Tn903) transposase rapidly processes the DNA at the IS(Tn) ends resulting in excision leading to direct insertion. Any delay in processing the 5’ cleavage at the transposon end which increases the half-life of the nicked intermediate would increase the probability of capturing a target DNA before processing leading to the formation of a DNA fork between donor and target DNA. This argument was based on knowledge of transposition of the Tn3 family of transposons and of the IS6 family of insertion sequences.
Direct (i.e non-replicative) transposition occurs by cleavage of both strands (3’ and 5’) at each transposon end to excise the transposon from its donor DNA and it is the 3’ ends with their 3’OH which are then used in a concerted attack of the target DNA (see: General_Information/Reaction_mechanisms; Fig. 23.1). For this reason, the strand with the 3’OH is called the transferred strand. Delayed cleavage of the 5’ or non-transferred strand, results in retention of the linkage between transposon and donor DNA and the creation of a branched structure by joint of the transferred strand to the target. This can act as the nucleus of a replication fork as occurs in transposition of the Tn3 and IS6 families of TE (see: Tn3_family/Replicative_transposition; Fig. Tn3.16; IS6_family/Mechanism:_the_state_of_play; Fig. IS6.8). This behavior has been demonstrated experimentally for transposon Tn7 (see: The TnsA-TnsB transposase complex and a switch between cut-and-paste and Replicative Transposition; Fig. Tn7.2C).
Bernardi and Bernardi [79] also investigated inter and intramolecular Tn903 transposition. They used a plasmid composed of the pSC101 plasmid and a segment of bacteriophage λ in which Tn903 had been inserted at one or the other plasmid/phage junction in either orientation. Their results were consistent with a replicative transposition component in Tn903 transposition but also indicated that one of the two flanking IS903 elements was preferentially used in inversion/deletion and cointegrate formation events. This behavior was dependent on the orientation of Tn903 in the donor plasmid and could reflect an effect of impinging transcription as has been found with Tn9/IS1.
Other IS903-flanked Compound Transposons
There are two additional documented IS903-based transposons: Tn603 [11], in which two directly repeated IS903 copies flank the same kanamycin resistance gene as in Tn903; and a less well documented structure lacking direct target repeats in which the pirA and pirB toxin genes are flanked by inverted copies of an IS similar to the IS903 group, ISVa2 ,in a structure called Pir Tn903 [87].
Tn9 (see also IS1 family)
Origin
Like Tn10 and Tn5, Tn9 (Fig. Tn9.1) was among the first compound transposons to be identified. Following the demonstration that phage P1 could acquire chloramphenicol resistance from a drug resistance plasmid [2], and its subsequent acquisition by phage[3] [4] isolated λ cam phage from a P1cam lysogen by thermal induction of λcI857. They showed that λcam phage were denser than the parental phage in CsCl gradients – indicating that they contained a DNA insertion and localized this by heteroduplex analysis. Acquisition appeared independent of the host rec and phage red recombination systems. They observed that insertions of identical size could occur at various places in the phage. In a series of articles in DNA Insertion Elements,Plasmids and Episomes (1977 ISBN0-87969-118-2), Rosner and Gottesman, Chow and Bukhari, MacHattie and Jakowski, and Yun and Vapnek all showed that similar sized chloramphenicol resistant insertions in phage P1 and Mu or phage λ basically using heteroduplex analysis. MacHattie and Jakowski used heteroduplexes with λ phage carrying insertions of IS1 to demonstrate that the cam insertions carried flanking directly repeated IS1 copies. Using restriction analysis and Southern hybridization of chloramphenicol resistant insertions in phage P1 and phage Mu [88]and concluded that Tn9 was flanked by two directly repeated copies of IS1.
Kondo and Mitsuhashi [2] observed that 5-20% of P1cam phage lose CamR on growth and this was accompanied by a decrease in density indicating loss of DNA. It was also noted that CamS phage retained part of the original insertion a length of DNA equivalent to the length of IS1. Similar instability was observed by MacHattie and Jakowski and by Gottesman and Rosner (1977 ISBN0-87969-118-2) using phage λ cam where a single plate lysate could include 10 - 70% CamS phage. Presumably these deletions occur by reciprocal recombination between the flanking IS1.
The complete Tn9 sequence is not available but has been constructed here by combining the sequence of the interstitial region containing the chloramphenicol transacetylase (CAT, CmR) gene [89] with two flanking directly repeated copies of IS1R (from ISfinder). It was shown to generate 9bp direct target repeats on insertion as do individual IS1 [90] (see: IS1 family). Transposition of IS1 appears to be able to occur through a number of pathways and this also seems to be the case for Tn9.
Amplification
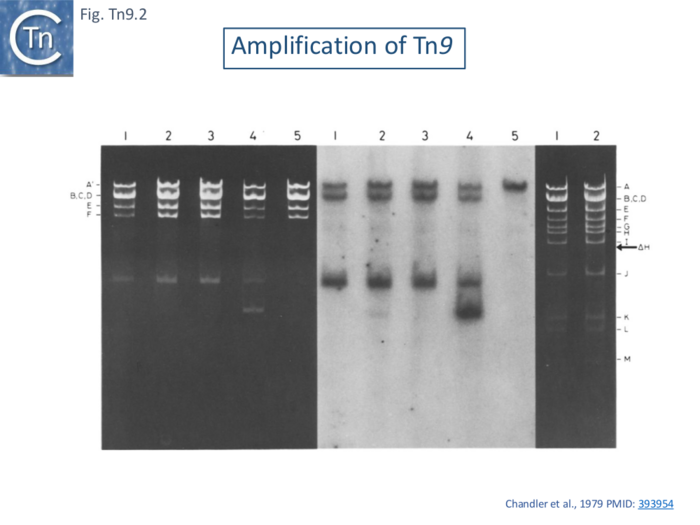
Tn9 was observed to form tandem dimers when harbored by small phage λ derivatives with smaller genomes capable of accommodating larger pieces of inserted DNA (MacHattie and Jakowski 1977 ISBN0-87969-118-2). This phenomenon was also observed when Tn9 was inserted into the resistance transfer factor (a derivative of R100.1 which lacks the major drug resistance gene cluster [91]; and the culture was grown with consecutive increases in the concentration of chloramphenicol in the medium up to 1mg/ml. Tn9 contains a single EcoRI site in the Cm gene and EcoRI digestion generated a fragment specific for Tn9 which can be detected simply in agarose gels (Fig. Tn9.2 A lane 4) or following a Southern blot using an IS1-specific probe (Fig. Tn9.2 B lane 4).
Two Transposition Pathways: Direct Transposition and Tn9-mediated Cointegrate Formation?
It was deduced by studying the frequency of transposition and cointegrate formation and the stability the transposition products that Tn9 transposition generates two types of mutually exclusive, end-products: either direct insertion into a recipient replicon (transposition), or fusion between donor and recipient replicons (cointegrate formation). This conclusion is based on the observations that, while Tn9-mediated cointegrates are very stable, they are formed at a rate lower than the transposition frequency. The ability of IS1 to promote these events is therefore also reflected in the behavior of Tn9.
The capacity of Tn9 and IS1 to generate cointegrates and to undergo transposition were measured in three ways: transposition from a λ::Tn9 prophage to an F-derived plasmid pOX38; cointegrate formation between pOX38::Tn9 and the non-conjugative plasmid, pBR322; and both cointegrate formation and transposition between the plasmid pBR322::Tn9 and pOX38 [92].
Three types of cointegrates were observed: replicon fusions using one or the other flanking IS1 and examples where the entire Tn9 had been used (Fig.Tn9.3). These structures were examined using restriction mapping of 40 individual clones using restriction mapping together with electron microscopy of 8 examples for confirmation. Of the 33 which used IS1A, 20 were simple fusions using IS1A (Fig.Tn9.3 B), 10 contained tandem copies of the pBR322 (Fig.Tn9.3 C) and 3 used IS1A but had multiple tandem pBR322 and Tn9 copies. Of the 4 which used IS1B, 3 were simple fusions (Fig.Tn9.3 D) and 1 carried a tandem pBR322 dimer (Fig.Tn9.3 E). Three examples had used the full length Tn9. Two were simple replicon fusions (Fig.Tn9.3 F) and one carried a pBR322 Tn9 dimer.
Subsequently, is was shown by identifying a number of transposition intermediates that IS1 can transpose in a number of different ways including direct transposition and cointegrate formation [93] (see: IS1 Family).
Similar relative cointegration frequencies of IS1A and IS1B were observed by Machida et al [94] using using a thermosensitive pSC101 plasmid, pHS1 [95] carrying Tn9 and plasmid colE1 as a target.
It was also observed that the individual flanking IS1 elements are more active in cointegrate formation than the complete Tn9 transposon underlining the “composite” nature of this transposon from a functional point of view.
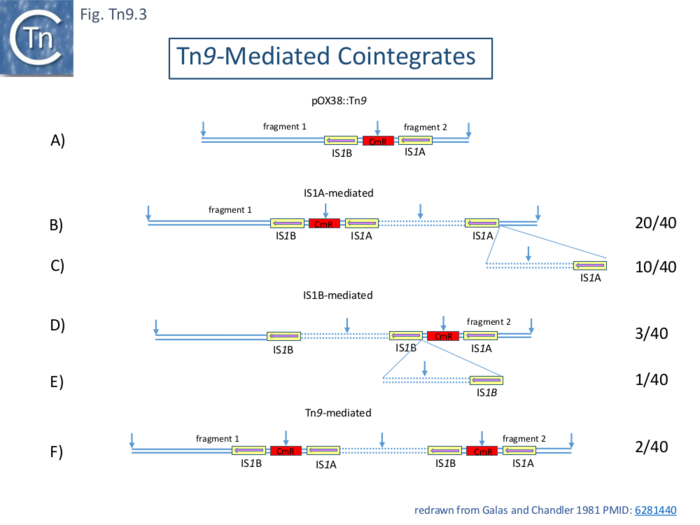
The Effect of Impinging Transcription
An additional observation was that, in both studies, the IS1 copy proximal to the cam gene promoter is more active than the other in forming cointegrates. This was initially ascribed to small differences in the sequences of the ends but subsequent analyses indicated that IS1 activity is inhibited by impinging transcription.
Two studies [97][98] explored this notion. The Ohtsubo lab [97] showed that removal of the cam promotor in Tn9 or the insertion of a phage T7 transcription terminator increased the ability of the downstream IS1 to form cointegrates again using pHS1 as a Tn9 donor. Moreover, it was demonstrated that in vitro transcripts of the cam gene in Tn9 read through into the downstream IS.
A different approach was used by Chandler and Galas [98] who came to the same conclusions in a continuing analysis of cointegrate formation. Here, Tn9 together with a short segment (132bp) of flanking DNA was cloned in different places and orientations into pBR322 and these were used as donors in a mating out assay using pOX38 as a target. The results showed that the IS1 copy proximal to a promoter exhibits lower activity than the distal copy and that deletion of the divergent promoters P1 and P2 reduces or eliminates this effect (Fig. Tn9.4)
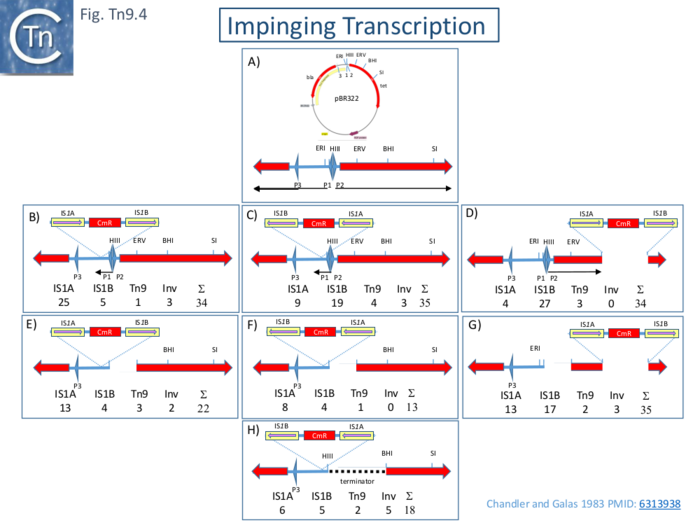
Tn9 Transposition frequency as a function of length: Transposition from a λ prophage
Early results from several laboratories had indicated that the frequency of transposition of various transposons flanked by ISI and carrying intervening DNA of different sizes and base composition varies over several orders of magnitude [4][99][100].
A systematic study of six IS1-flanked transposons ranging in length from 2.64 to approximately 10.5 kb inserted into a phage prophage was undertaken using both pOX38 and an R100.1 RTF derivative as targets.
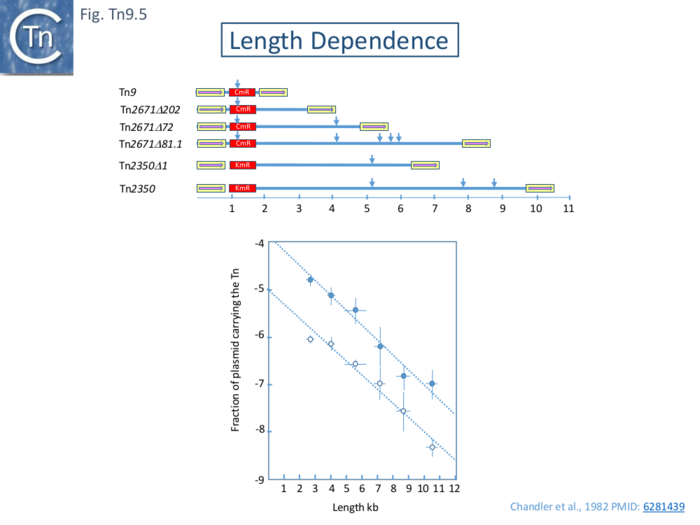
The results (Fig. Tn9.5) clearly show that with both target plasmids, there is a logarithmic decrease in transposition activity over this size range and is reduced by approximately 50% with each increase of 1kb of DNA. This is somewhat higher than was found with Tn10 (Fig. Tn10.5). One suggestion which has yet to be rigorously demonstrated was that transposition occurs by a progressive tracking mechanism with a constant probability of abortion per unit DNA length. Another would be that the probability of the two IS1 outside ends to synapse to form a transpososome simply decreases with increasing distance between the two.
Other IS1-flanked Compound Transposons
There are three principal IS1 transposons whose transposition properties have been observed. One of these, Tn1681, carries a heat stable (STI) toxin gene from Escherichia coli and formed stem loop structures in heteroduplex electron microscopy studies [9] exhibit a stem and loop structure with a stem of approximately 800bp suggesting terminal inverted copies of IS1. This was confirmed by its DNA sequence [102]. Little attention has been directed to studying Tn1681 transposition although the transposon has been detected in other genetic environments (refs).
Another IS1-flanked transposon, Tn2350, was isolated by transposition into a λ phage from the multiple antibiotic resistance plasmid, R1drd19[100]. Tn2350 is approximately 6kb in length and specifies resistance to kanamycin. It is flanked by directly repeated IS1 copies identified by Southern hybridization, e.m. heteroduplex analysis and restriction mapping. Like the IS1-flanked r-det of plasmid R100.1, it can be observed as a circular form carrying a single IS1 copy in dnaAts strains integratively suppressed by insertion of R1drd19 presumably formed by recombination between the two flanking IS1. However, unlike the xkb r-det, the isolated circles can be maintained when transformed into a recA- E. coli strain [103]. However, this is not the result of replication but of extremely efficient and reversible recombination with the xer site in the E.coli chromosome [104].
There are many examples of various genes flanked by IS1 copies in both chromosome and plasmid DNA sequences in the public databases although these have not been investigated either for flanking direct target repeats or for their capacity to transpose.
Tn6330: an IS30 family Transposon
IS30 family members transpose by the copy-out-paste-in mechanism producing circular transposition intermediates with abutting IS ends. An important IS30-based transposon, Tn6330, has been identified as responsible for the worldwide transmission of resistance to one of the last resort antibiotic, colistin.
Origin
Tn6330, encodes the mcr-1 (colistin resistance) gene flanked by copies of the IS30 family member ISApl1. A common 2,607-bp DNA segment, containing the mcr-1 gene together with a truncated copy of a gene called pap2, was identified in mcr-1-containing sequences that in many cases was flanked on one or both ends by ISApl1 [105] and it was proposed mcr-1 was transmitted by an ISApl1 composite transposon, later called Tn6330 [106] , which has, in some cases, subsequently lost one or both ISApl1 copies. Like members of the IS30 family itself, circular copies of the entire transposon, presumably transposition intermediates, have been observed [107].
Birth of Tn6330?
Further studies identified the potential source of the mcr-1 and pap2 DNA segment in the chromosome of Moraxella sp. MSG13-C03 and suggested that Tn6330 was created by insertion of ISApl1 copies upstream and downstream (Fig. IS30.7). It was noted that these regions were rich in poly-A sequence, a preferred ISApl1 insertion site [105].
The Demise of Tn6330: Disguising the transposon source of horizontally transmitted genes.
Analysis of the collection of mcr-1 genes also revealed the presence of small remnants (scars) of a few base pairs of ISApl1. This led to the idea that the flanking IS were unstable and relatively rapidly eliminated by deletion.
A model, based on DNA strand switching during the copy-out reaction which precedes the paste-in event terminating transposition, was proposed to explain the loss of flanking ISApl1 copies (Fig. IS30.10; [108]). This received strong support [109] by the identification of gene [110][111] encoding a so-called prim-pol enzyme [112] which would permit strand switching at microhomologies and also provide a primase function resulting in excision of certain types of IS [113][114] (see IS3_family/Excision:_A_dedicated_enzyme). The enzyme was shown to enhance excision of IS which transpose by the copy-out-paste-in mechanism but not IS with other transposition mechanisms (Fig. IS3.25; Fig. IS3.26).
Clearly, relatively high instability of Tn6330 and removal of its flanking IS would mask the fact that mcr-1 had been horizontally transferred by a compound transposon. Since the prim-pol gene is distributed in many enteric bacteria, it seems possible that a number of “horizontally” transmitted genes have been transferred in this way and might also contain small scars from their ancestral flanking IS.
IS256 family Transposons
There are a number of compound transposons based on IS256 family members which are clinically important.
Tn4001
Transposon Tn4001, which encodes the aacA-aphD genes specifying resistance to gentamicin, tobramycin and kanamycin flanked by inverted copies of IS256, Tn4001 was originally isolated from Staphylococcus aureus and shown to transpose from a plasmid into the S. aureus chromosome [115] and is 4566 bp long [116]. It has proved useful as a tool for genetic analysis of a number of gram-positive bacteria [117][118][119][120][121][122].
Excised circular copies of both the flanking IS256 and the entire transposon have been identified in Streptococcus pneumoniae [123] as might be expected for transposition using a copy-out-paste-in transposition pathway. The interstitial base pairs between the two Tn IRs in these circles is five of six base pairs but insertion, like that of IS256 itself, generates an 8bp DR. This presumably reflects a difference in the architecture of the transpososome formed during excision and integration.
Three probable Tn4001 homologues, based on detailed restriction maps, are Tn4031 from S. epidermidis [124], Tn5281 from Enterococcus faecalis [125] and Tn3706 from Streptococcus agalactiae B128 [126]. Another related transposon, Tn5384, identified in E. faecalis CH116 confers resistance to erythromycin and to high levels of gentamicin [127]. It is 26 kb long and appears to include a transposon very similar to Tn4001 with its inverted IS256 copies flanking the aacA-aphD genes together with a large downstream DNA segment carrying an erythromycin and mercuric chloride resistance genes [128] and terminating with a third IS256 copy. Tn5384 uses one IS256 copy from Tn4001 together with the third IS256 copy present in direct repeat. Tn5384 insertions into the enterococcal plasmid pLRM1 were flanked by 8- or 9-bp direct target duplications.
Tn1547
A 64-kb vanB vancomycin-resistance (VmR) was identified by transposition from the Enterococcus faecalis BM4281 chromosome into the hemolysin (Hly) plasmid, pIP964 [129]. Although the VmR genes carried by this transposon, Tn1547, are flanked by two directly repeated but non-identical IS, which are similar enough to permit their concerted use in transposition. One is an IS256 copy (with two nucleotide transitions in the transposase gene) while the other, called IS16, is longer (1466 instead of 1324bp). Its transposase gene was 61% identical to that of IS256 with 58% identity between the 390 aa IS256 and the 395 aa IS16 transposases. Importantly, the 18/26 bp IR were closely related (23/26bp). Tn1547 insertion results in an 8 bp target duplication.
IS6 family and Pseudocompound Transposons
The term pseudocompound transposons (Galas and Chandler 1989 ISBN: 978-1555810054) was coined to describe transposons which are based on IS6 family members. The flanking IS are always in direct repeat because IS6 family members transpose via the formation of cointegrates followed by a resolution by recombination (Fig. IS6.8). This is treated in detail in the chapter concerning the IS6 family. Members of the IS6 family, in particular, IS26, play an important role in plasmid rearrangements and fusions and in gene amplification. There are many examples of genes flanked by directly repeated IS6 family copies. However, although direct transposition of IS6 members generate target repeats of 8bp, intra molecular transpositions which lead to deletions and inversions, do not generate the flanking repeats (Fig. IS6.9). To determine whether genes flanked by direct IS6 family copies are indeed part of a compound transposon in the absence of direct target repeats, it is necessary to identify the same structures in different genetic environments. Many of the IS6 family-based compound transposons listed in TnCentral which have been assigned Tn identification numbers have not fulfilled either criterion.
Integrative and Conjugative Elements (ICE) Driven by DDE Transposases.
Integrative Conjugative Transposons (ICE) are transposable elements which reside in the host chromosome but are capable of excising as a circular copy and can then undergo conjugation into other hosts. While many are autonomous, some require functions which can be supplied in trans from another ICE in the same cell. They commonly excise and integrate using sequence-specific recombinases [130][131]. However, several ICE have been identified which, instead of encoding site-specific recombination machinery, carry DDE transposases [132][133][134].
TnGBS1 and TnGBS2
These were initially identified by genome comparisons of Streptococcus agalactiae which revealed a composite organization composed of a conserved backbone with many (11–14) large variable regions resembling genomic islands [135]. A later study of eight S. agalactiae genomes identified 35 of these regions including a number of putative ICE. Circular forms of five of these were detected [136]. One chromosomal ICE (originally called X) was also observed to mobilize chromosomal DNA in a similar way to Hfr conjugative transfer [137]. In one strain (NEM316) there were three copies of one putative ICE, pNEM316-1 [137] whose junctions with the backbone DNA exhibited identical 25bp imperfect inverted repeats and 9-10 bp flanking direct repeats. In two cases, an “empty” chromosome sequence was identified demonstrating that the DRs were generated from the target DNA as a consequence of insertion. A circular species was also identified in which the IR were abutted and separated by 8bp whose sequence could not be determined, presumably because the circles were derived from a number of inserted elements with identical IRs [138]. The X (33.5 kbp) and pNEM316-1 (47 kbp) elements were subsequently renamed TnGBS2 and TnGBS1 respectively and were found to contain a DDE transposase related to the eukaryotic Mutator-like transposases (MULT) at their right ends [132](Fig. DDE ICE.1) Further studies showed that this was very closely related to the prokaryotic ISLre2. They were also shown to have flanking IRs.
To explore the relationship between ICE and the MULT group, genes were retrieved by cascade PSI-Blast using as initial query the transposase of TnGBS2 (see IS256_family/The_IS256_cluster [134]. These fell into 5 groups, MULT 1-5, based on their transposase, sequences (Fig. IS256.1), their active sites (Fig. IS256.2) and on their left and right ends. Maps of a sample of DDE ICE together with there is relatives are shown in Fig. DDE ICE.2.
Bibliography
- ↑ Mobile DNA. American Society for Microbiology. 1989. Galas and Chandler 1989 ISBN: 978-1555810054
- ↑ 2.0 2.1 2.2 <pubmed>14234780</pubmed>
- ↑ 3.0 3.1 <pubmed>4576341</pubmed>
- ↑ 4.0 4.1 4.2 <pubmed>1061090</pubmed>
- ↑ 5.0 5.1 5.2 <pubmed>1059152</pubmed>
- ↑ <pubmed> 1092669</pubmed>
- ↑ 7.0 7.1 7.2 7.3 7.4 <pubmed> 1102715</pubmed>
- ↑ <pubmed> 383689</pubmed>
- ↑ 9.0 9.1 <pubmed>368646</pubmed>
- ↑ 10.0 10.1 <pubmed>723960</pubmed>
- ↑ 11.0 11.1 <pubmed>2819910</pubmed>
- ↑ <pubmed>3033719</pubmed>
- ↑ <pubmed>26104553</pubmed>
- ↑ <pubmed>1104574</pubmed>
- ↑ <pubmed>792710</pubmed>
- ↑ <pubmed>365678</pubmed>
- ↑ 17.0 17.1 <pubmed>378398</pubmed>
- ↑ <pubmed>6279310</pubmed>
- ↑ 19.0 19.1 19.2 <pubmed>231729</pubmed>
- ↑ 20.0 20.1 <pubmed>455447</pubmed>
- ↑ <pubmed>10781570</pubmed>
- ↑ <pubmed> 26104553</pubmed>
- ↑ <pubmed>4909470</pubmed>
- ↑ <pubmed>337110</pubmed>
- ↑ <pubmed>6289329</pubmed>
- ↑ <pubmed>772162</pubmed>
- ↑ <pubmed>340912</pubmed>
- ↑ 28.0 28.1 28.2 <pubmed>1648003</pubmed>
- ↑ <pubmed>158473</pubmed>
- ↑ 30.0 30.1 <pubmed>370414</pubmed>
- ↑ <pubmed> 340912</pubmed>
- ↑ <pubmed>6091910</pubmed>
- ↑ <pubmed>2553269</pubmed>
- ↑ <pubmed>455446</pubmed>
- ↑ <pubmed>6311438</pubmed>
- ↑ <pubmed>1691097</pubmed>
- ↑ <pubmed>25649688</pubmed>
- ↑ <pubmed> 455446</pubmed>
- ↑ 39.0 39.1 39.2 <pubmed>6260376</pubmed>
- ↑ <pubmed>6264444</pubmed>
- ↑ <pubmed>3038673</pubmed>
- ↑ <pubmed>6287993</pubmed>
- ↑ 43.0 43.1 <pubmed>2981756</pubmed>
- ↑ <pubmed>6322169</pubmed>
- ↑ <pubmed>1648666</pubmed>
- ↑ <pubmed>9639701</pubmed>
- ↑ <pubmed>160562</pubmd>
- ↑ <pubmed>7679474</pubmed>
- ↑ <pubmed>2167449</pubmed>
- ↑ <pubmed>7526163</pubmed>
- ↑ <pubmed>7523934</pubmed>
- ↑ <pubmed>9403037</pubmed>
- ↑ <pubmed>10102036</pubmed>
- ↑ <pubmed>328481</pubmed>
- ↑ <pubmed>381840</pubmed>
- ↑ <pubmed>6260375</pubmed>
- ↑ <pubmed> 6299577</pubmed>
- ↑ 58.0 58.1 <pubmed> 2998922</pubmed>
- ↑ <pubmed>6281439</pubmed>
- ↑ 60.0 60.1 60.2 <pubmed>6248868</pubmed>
- ↑ <pubmed>12511513</pubmed>
- ↑ 62.0 62.1 <pubmed>6305767</pubmed>
- ↑ <pubmed>2872578</pubmed>
- ↑ <pubmed>388418</pubmed>
- ↑ <pubmed>14310189</pubmed>
- ↑ <pubmed>345091</pubmed>
- ↑ 67.0 67.1 67.2 <pubmed>6244898</pubmed>
- ↑ <pubmed>390162</pubmed>
- ↑ <pubmed>6260374</pubmed>
- ↑ <pubmed>6283538</pubmed>
- ↑ <pubmed>6247321</pubmed>
- ↑ <pubmed>770430</pubmed>
- ↑ 73.0 73.1 <pubmed>344143</pubmed>
- ↑ <pubmed>4580676</pubmed>
- ↑ 75.0 75.1 <pubmed>6261245</pubmed>
- ↑ <pubmed>6270337</pubmed>
- ↑ 77.0 77.1 <pubmed>6314502</pubmed>
- ↑ 78.0 78.1 <pubmed>6099240</pubmed>
- ↑ 79.0 79.1 <pubmed>1646385</pubmed>
- ↑ 80.0 80.1 <pubmed>11387225</pubmed>
- ↑ <pubmed>6248736</pubmed>
- ↑ <pubmed>6267289</pubmed>
- ↑ 83.0 83.1 <pubmed>7029308</pubmed>
- ↑ <pubmed>6292435</pubmed>
- ↑ <pubmed>6289304</pubmed>
- ↑ <pubmed>6298776</pubmed>
- ↑ <pubmed>28169338</pubmed>
- ↑ <pubmed>365686</pubmed>
- ↑ <pubmed>390403</pubmed>
- ↑ <pubmed>365355</pubmed>
- ↑ <pubmed>393954</pubmed>
- ↑ <pubmed>6281440</pubmed>
- ↑ <pubmed>7489730</pubmed>
- ↑ <pubmed>6281761</pubmed>
- ↑ <pubmed>783149</pubmed>
- ↑ <pubmed> 6281440</pubmed>
- ↑ 97.0 97.1 <pubmed>6309405</pubmed>
- ↑ 98.0 98.1 <pubmed>6313938</pubmed>
- ↑ 99.0 99.1 <pubmed>385224</pubmed>
- ↑ 100.0 100.1 100.2 <pubmed>6255291</pubmed>
- ↑ <pubmed> 390403</pubmed>
- ↑ <pubmed>6254008</pubmed>
- ↑ <pubmed>6284717</pubmed>
- ↑ <pubmed>1931823</pubmed>
- ↑ 105.0 105.1 <pubmed>27620479</pubmed>
- ↑ <pubmed>30927434</pubmed>
- ↑ <pubmed>30723461</pubmed>
- ↑ <pubmed>29440577</pubmed>
- ↑ <pubmed>37574851</pubmed>
- ↑ <pubmed>10698782</pubmed>
- ↑ <pubmed>16233651</pubmed>
- ↑ <pubmed>36715333</pubmed>
- ↑ <pubmed>24334665</pubmed>
- ↑ <pubmed>21224843</pubmed>
- ↑ <pubmed>6323927</pubmed>
- ↑ <pubmed>2553542</pubmed>
- ↑ <pubmed>7597109</pubmed>
- ↑ <pubmed>10894746</pubmed>
- ↑ <pubmd>10087215</pubmed>
- ↑ <pubmed>9073584</pubmed>
- ↑ <pubmed>10903438</pubmed>
- ↑ <pubmed>8106329</pubmed>
- ↑ <pubmed>11751820</pubmed>
- ↑ <pubmed>2552907</pubmed>
- ↑ <pubmed>1656854</pubmed>
- ↑ <pubmed>8723445</pubmed>
- ↑ <pubmed>7625803</pubmed>
- ↑ <pubmed>9303373</pubmed>
- ↑ <pubmed>8654967</pubmed>
- ↑ <pubmed>26473380</pubmed>
- ↑ <pubmed>20601965</pubmed>
- ↑ 132.0 132.1 <pubmed>19183283</pubmed>
- ↑ <pubmed>23435978</pubmed>
- ↑ 134.0 134.1 <pubmed>24418649</pubmed>
- ↑ <pubmed>16172379</pubmed>
- ↑ <pubmed>18708498</pubmed>
- ↑ 137.0 137.1 <pubmed>18832470</pubmed>
- ↑ <pubmed>12354221</pubmed>