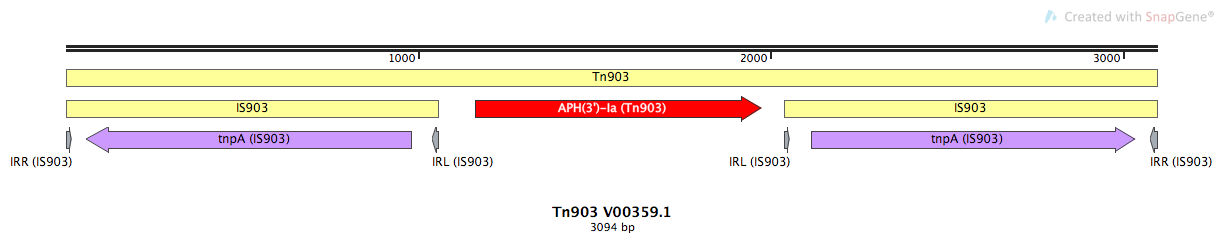
Mobile element type:
Transposon
Name:
Tn903
Synonyms:
Accession:
Tn903-V00359.1
Family:
Compound Transposon
Group:
IS903
First isolate:
Yes
Partial:
ND
Evidence of transposition:
Yes
Host Organism:
Escherichia coli
Date of Isolation:
1978
Country:
Molecular Source:
plasmid R6-5
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| tnpA | IS903 | 57-980 | - | Transposase | None |
| APH(3')-Ia (ARO:3002641) | Tn903 | 1162-1977 | + | Passenger Gene | Antibiotic Resistance |
| tnpA | IS903 | 2115-3038 | + | Transposase | None |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | IS903 | 307 | 57-980 | - |
|---|
| Class: | Transposase | ||||||||||
| Function: | GO molecular function: DNA binding, transposase activity. | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MAKQKFKITN WSTYNKALIN RGSLTFWLDD GAIQAWYESA TPSSRGRPQR YSDLAITTVL VIKRVFRLTL RAAQGFIDSI FTLMNVPLRC PDYSCVSRRA KSVNISFKTF TRGEIAHLVI DSTGLKVFGE GEWKVKKHGQ ERRRIWRKLH LAVDSKTHEI ICADLSLNNV TDSEAFPGLI RQTHRKIRAA SADGAYDTRL CHDELRRKKI SALIPPRKGA GYWPGEYADR NRAVANQRMT GSNARWKWTT DYNRRSIAET AMYRVKQLFG GSLTLRDYDG QVAEAMALVR ALNKMTKAGM PESVRIA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | APH(3')-Ia (ARO:3002641) | APH(3')-Ia | Tn903 | 271 | 1162-1977 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | APH(3') (ARO:3000126) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | strict match to reference sequence for ARO:3002641 (bitscore: 560)||Synonyms: aphA-1,apha1-1AB, APH(3')-Ic, apha7 | ||||||||||
| Protein Sequence: | MSHIQRETSC SRPRLNSNMD ADLYGYKWAR DNVGQSGATI YRLYGKPDAP ELFLKHGKGS VANDVTDEMV RLNWLTEFMP LPTIKHFIRT PDDAWLLTTA IPGKTAFQVL EEYPDSGENI VDALAVFLRR LHSIPVCNCP FNSDRVFRLA QAQSRMNNGL VDASDFDDER NGWPVEQVWK EMHKLLPFSP DSVVTHGDFS LDNLIFDEGK LIGCIDVGRV GIADRYQDLA ILWNCLGEFS PSLQKRLFQK YGIDNPDMNK LQFHLMLDEF F |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | IS903 | 307 | 2115-3038 | + |
|---|
| Class: | Transposase | ||||||||||
| Function: | GO molecular function: DNA binding, transposase activity. | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MAKQKFKITN WSTYNKALIN RGSLTFWLDD GAIQAWYESA TPSSRGRPQR YSDLAITTVL VIKRVFRLTL RAAQGFIDSI FTLMNVPLRC PDYSCVSRRA KSVNISFKTF TRGEIAHLVI DSTGLKVFGE GEWKVKKHGQ ERRRIWRKLH LAVDSKTHEI ICADLSLNNV TDSEAFPGLI RQTHRKIRAA SADGAYDTRL CHDELRRKKI SALIPPRKGA GYWPGEYADR NRAVANQRMT GSNARWKWTT DYNRRSIAET AMYRVKQLFG GSLTLRDYDG QVAEAMALVR ALNKMTKAGM PESVRIA |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| IS903-V00359.1 | IS903 | insertion sequence | 1-1057 | + | 1057 |
| IS903-V00359.1 | IS903 | insertion sequence | 2038-3094 | + | 1057 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRR | IS903 | 1-18 | + | 18 | GGCTTTGTTG AATAAATC |
| IRL | IS903 | 1040-1057 | - | 18 | GGCTTTGTTG AATAAATC |
| IRL | IS903 | 2038-2055 | + | 18 | GGCTTTGTTG AATAAATC |
| IRR | IS903 | 3077-3094 | - | 18 | GGCTTTGTTG AATAAATC |
1. Hershfield V, Boyer HW, Chow L, Helinski DR.
Characterization of a mini-ColC1 plasmid.. J Bacteriol. 1976 Apr;126(1):447-53. doi: 10.1128/jb.126.1.447-453.1976. PubMed ID:
770430
.
2. Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903.. J Mol Biol. 1981 Apr 5;147(2):217-26. doi: 10.1016/0022-2836(81)90438-1. PubMed ID: 6270337 .
3. Nomura N, Yamagishi H, Oka A. Isolation and characterization of transducing coliphage fd carrying a kanamycin resistance gene.. Gene. 1978 Feb;3(1):39-51. doi: 10.1016/0378-1119(78)90006-9. PubMed ID: 344143 .
4. Grindley ND, Joyce CM. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903.. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7176-80. doi: 10.1073/pnas.77.12.7176. PubMed ID: 6261245 .
5. Swingle B, O'Carroll M, Haniford D, Derbyshire KM. The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10.. Mol Microbiol. 2004 May;52(4):1055-67. doi: 10.1111/j.1365-2958.2004.04051.x. PubMed ID: 15130124 .
6. Twiss E, Coros AM, Tavakoli NP, Derbyshire KM. Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress.. Mol Microbiol. 2005 Sep;57(6):1593-607. doi: 10.1111/j.1365-2958.2005.04794.x. PubMed ID: 16135227 .
2. Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903.. J Mol Biol. 1981 Apr 5;147(2):217-26. doi: 10.1016/0022-2836(81)90438-1. PubMed ID: 6270337 .
3. Nomura N, Yamagishi H, Oka A. Isolation and characterization of transducing coliphage fd carrying a kanamycin resistance gene.. Gene. 1978 Feb;3(1):39-51. doi: 10.1016/0378-1119(78)90006-9. PubMed ID: 344143 .
4. Grindley ND, Joyce CM. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903.. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7176-80. doi: 10.1073/pnas.77.12.7176. PubMed ID: 6261245 .
5. Swingle B, O'Carroll M, Haniford D, Derbyshire KM. The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10.. Mol Microbiol. 2004 May;52(4):1055-67. doi: 10.1111/j.1365-2958.2004.04051.x. PubMed ID: 15130124 .
6. Twiss E, Coros AM, Tavakoli NP, Derbyshire KM. Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress.. Mol Microbiol. 2005 Sep;57(6):1593-607. doi: 10.1111/j.1365-2958.2005.04794.x. PubMed ID: 16135227 .