Difference between revisions of "General Information"
Jump to navigation
Jump to search
| (36 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
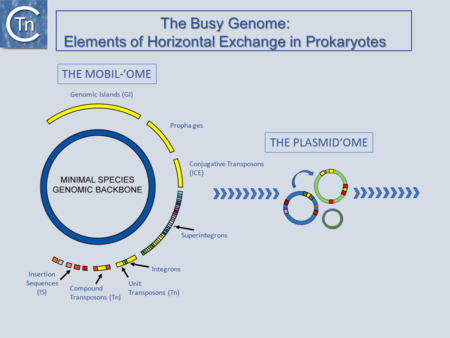
| − | ; 1. [[General Information/Overview|Overview]] | + | [[Image:fig1.png|thumb|450x450px|[[General Information/Overview|'''Fig.1.1. The Busy Genome:''']] Elements of Horizontal Exchange. The genome backbone, which includes housekeeping genes, is shown as the inner circle (blue). The "mobilome" is shown in the outer circle. This includes a number of different types of MGE both intercellular (some genomic islands, prophages, and conjugative transposons) and intracellular (Insertion sequences, compound and unit transposons, integrons, and super integrons). An important class of intercellular MGE, the plasmids, act as transposon vectors and facilitate TE movement within the plasmidome.|alt=|border]] |
| + | {| class="wikitable" | ||
| + | |+ | ||
| + | | width="680pt" | | ||
| + | ;1. [[General Information/Overview|Overview]] | ||
| − | ; 2. [[General Information/IS History| | + | ;2. [[General Information/IS History|Insertion Sequence History and Early Transposition Models]] |
| − | ; 3. [[General Information/What Is an IS?|What Is an IS?]] | + | ;3. [[General Information/What Is an IS?|What Is an IS?]] |
| − | ; 4. [[General Information/ISfinder and the Growing Number of IS|ISfinder and the Growing Number of IS]] | + | ;4. [[General Information/ISfinder and the Growing Number of IS|ISfinder and the Growing Number of IS]] |
| − | ; 5. [[General Information/IS Identification|IS Identification]] | + | ;5. [[General Information/IS Identification|IS Identification, nomenclature and naming attribution]] |
| − | ; 6. [[General Information/IS Distribution|IS Distribution]] | + | ;6. [[General Information/IS Distribution|IS Distribution]] |
| − | ; 7. [[General Information/Major Groups are Defined by the Type of Transposase They Use|Major Groups are Defined by the Type of Transposase They Use]] | + | ;7. [[General Information/Major Groups are Defined by the Type of Transposase They Use|Major Groups are Defined by the Type of Transposase They Use]] |
| − | ; 8. [[General Information/Fuzzy Borders|Fuzzy Borders]] | + | ;8. [[General Information/Fuzzy Borders|Fuzzy Borders]] |
| − | ; 9. [[General Information/tIS - IS and relatives with passenger genes|tIS - IS and relatives with passenger genes]] | + | ;9. [[General Information/tIS - IS and relatives with passenger genes|tIS - IS and relatives with passenger genes]] |
| − | ; 10. [[General Information/ IS derivatives of Tn3 family transposons| IS derivatives of | + | ;10. [[General Information/ IS derivatives of Tn3 family transposons| IS derivatives of Tn''3'' family transposons]] |
| − | ; 11. [[General Information/IS related to Integrative Conjugative Elements (ICEs)|IS related to Integrative Conjugative Elements (ICEs)]] | + | ;11. [[General Information/IS related to Integrative Conjugative Elements (ICEs)|IS related to Integrative Conjugative Elements (ICEs)]] |
| − | ; 12. [[General Information/IS91 and ISCR| | + | ;12. [[General Information/IS91 and ISCR|IS''91'' and IS''CR'' families]] |
| − | ; 13. [[General Information/Non-autonomous IS derivatives|Non-autonomous IS derivatives]] | + | ;13. [[General Information/Non-autonomous IS derivatives|Non-autonomous IS derivatives]] |
| − | ; 14. [[General Information/ Relationship Between IS and Eukaryotic TE| Relationship Between IS and Eukaryotic TE]] | + | ;14. [[General Information/ Relationship Between IS and Eukaryotic TE| Relationship Between IS and Eukaryotic TE]] |
| − | ; 15. [[General Information/ Impact of IS on Genome Evolution - The Importance of Time Scale| Impact of IS on Genome Evolution - The Importance of Time Scale]] | + | ;15. [[General Information/ Impact of IS on Genome Evolution - The Importance of Time Scale| Impact of IS on Genome Evolution - The Importance of Time Scale]] |
| − | ; 16. [[General Information/Target Choice|Target Choice]] | + | ;16. [[General Information/Target Choice|Target Choice]] |
| − | ; 17. [[General Information/Influence of transposition mechanisms on genome impact|Influence of transposition mechanisms on genome impact]] | + | ;17. [[General Information/Influence of transposition mechanisms on genome impact|Influence of transposition mechanisms on genome impact]] |
| − | ; 18. [[General Information/IS and Gene Expression|IS and Gene Expression]] | + | ;18. [[General Information/IS and Gene Expression|IS and Gene Expression]] |
| − | ; 19. [[General Information/IS Organization|IS Organization]] | + | ;19. [[General Information/IS Organization|IS Organization]] |
| − | ; 20. [[General Information/Control of transposition activity|Control of transposition activity]] | + | ;20. [[General Information/Control of transposition activity|Control of transposition activity]] |
| − | ; 21. [[General Information/Transposase expression and activity|Transposase expression and activity]] | + | ;21. [[General Information/Transposase expression and activity|Transposase expression and activity]] |
| − | ; 22. [[General Information/ | + | ;22. [[General Information/Reaction mechanisms|Reaction mechanisms]] |
| − | ; 23. [[General Information/ | + | ;23. [[General Information/The casposases|The Casposases]] |
| − | ; | + | |} |
| + | |||
| + | ; | ||
| + | |||
| + | <br/> | ||
| + | <br/> | ||
| + | |||
| + | {{TnPedia}} | ||
| + | |||
| + | <htmlet nocache="yes" >foobar</htmlet> | ||
Latest revision as of 12:15, 5 December 2022

Fig.1.1. The Busy Genome: Elements of Horizontal Exchange. The genome backbone, which includes housekeeping genes, is shown as the inner circle (blue). The "mobilome" is shown in the outer circle. This includes a number of different types of MGE both intercellular (some genomic islands, prophages, and conjugative transposons) and intracellular (Insertion sequences, compound and unit transposons, integrons, and super integrons). An important class of intercellular MGE, the plasmids, act as transposon vectors and facilitate TE movement within the plasmidome.
|