Transposons families/Tn7 family
Historical
Plasmid R483 was first suspected to carry a trimethoprim and streptomycin resistance transposon because these plasmid determinants became genetically linked to and transferred by a second plasmid, R144, in the host strain [1]. That the two resistance genes were part of a transposable element was confirmed by showing that they could be transposed from the incI→ plasmid, R483, to plasmids of other incompatibility groups such as R391(IncJ), R7K(IncW), and RP4(IncP), which all acquired a DNA segment of about 9 x 106 daltons as judged by sucrose density centrifugation [2]. Remarkably, in contrast to other transposons at the time, transposition into the host E. coli chromosome was also observed to a single locus between dnaA and ilv [2]. The authors pointed out that insertion did not suppress a dnaAts phenotype indicating that it did not involve the entire R483 plasmid. This is because an integrated plasmid was known to suppress dnaAts thermosensitivity by providing an alternative (integrated plasmid-driven) origin of replication for the chromosome [3][4][5], a phenomenon known as integrative suppression [6]. The chromosomally located resistance determinant could undergo subsequent transposition to another plasmid target in a recA independent manner. The transposable resistance determinant was given the name TnC [2] which later became Tn7 [7][8].
The Unique Chromosome insertion site attTn7
The nature of the specific chromosomal insertion site was investigated further [9]: by conjugation, where it was shown to be tightly genetically linked to the chromosomal markers glmS and ilv; by restriction enzyme mapping and hybridization, where it was shown to have integrated into the chromosome at the same unique position in a number of independent “transposants”; and by cloning the chromosome region into a suitable vector plasmid where it was shown that the chromosomal fragment acted as an efficient Tn7 transposition target when cloned in either orientation and, moreover, that Tn7 insertion always occurred in the same orientation with respect to the cloned fragment.
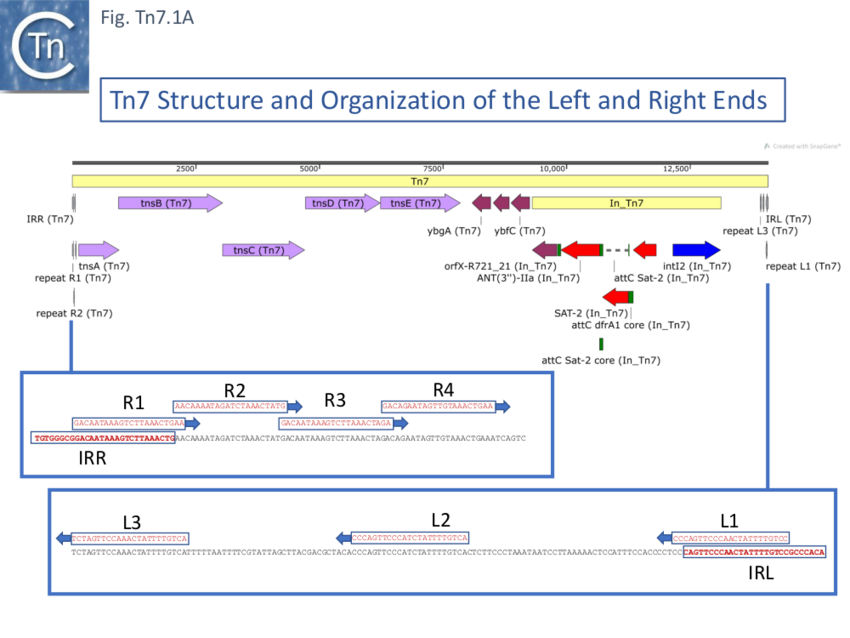
Further analysis of the chromosomal insertion site (which the authors called attTn7 in analogy to bacteriophage attB sites) by DNA sequence analysis [10] identified the junction between the target and each Tn7 end and revealed that insertion created a 5bp direct target repeat and that the ends exhibited short terminal inverted repeats but contained, 22bp repeated sequences over a longer distance which were asymmetric at each end (Fig. Tn7.1A) (see also [11]). These were contiguous in the right end but were separated at the left end [10]. It was suggested that this asymmetry may play a role in the unique insertion orientation of Tn7 in the attachment site. Note that for historical reasons, in contrast to other transposons, the left and right transposon ends are inversed for Tn7. (That is: the end proximal to the transposition genes is labelled Tn7R whereas in most other transposition systems the convention is that the transposase proximal end is defined as IRL). Later studies revealed that 199 bp of Tn7R and 166 bp of Tn7L were sufficient for transposition activity when transposition functions were provided in trans [12].
Bibliography
- ↑
- ↑ 2.0 2.1 2.2 Barth PT, Datta N, Hedges RW, Grinter NJ . Transposition of a deoxyribonucleic acid sequence encoding trimethoprim and streptomycin resistances from R483 to other replicons. - J Bacteriol: 1976 Mar, 125(3);800-10 [PubMed:767328] [DOI]
- ↑
- ↑ Bird RE, Chandler M, Caro L . Suppression of an Escherichia coli dnaA mutation by the integrated R factor R.100.1: Change of chromosome replication origin in synchronized cultures. - J Bacteriol: 1976 Jun, 126(3);1215-23 [PubMed:780344] [DOI]
- ↑ Chandler M, Silver L, Roth Y, Caro L . Chromosome replication in an Hfr strain of Escherichia coli. - J Mol Biol: 1976 Jun 25, 104(2);517-23 [PubMed:781291] [DOI]
- ↑ Nishimura Y, Caro L, Berg CM, Hirota Y . Chromosome replication in Escherichia coli. IV. Control of chromosome replication and cell division by an integrated episome. - J Mol Biol: 1971 Feb 14, 55(3);441-56 [PubMed:4927942] [DOI]
- ↑ Lederberg EM. Plasmid reference center registry of transposon (Tn) allocations through July 1981. Gene. 1981;16:59–61.
- ↑ Campbell A, Berg DE, Botstein D, Lederberg EM, Novick RP, Starlinger P, Szybalski W. Nomenclature of transposable elements in prokaryotes. Gene. 1979;5:197–206.
- ↑
- ↑ 10.0 10.1
- ↑
- ↑