Difference between revisions of "IS Families/ISAzo13 family"
Jump to navigation
Jump to search
(Created page with "This family, represented by 37 members in ISfinder emerged from the ISNCY orphan group. It is based on both Tpase and IR sequence similarities (Fig. ISAzo13.1). Insertion gene...") |
|||
| (11 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | '''<big>T</big>'''his family, represented by 37 members in [https://isfinder.biotoul.fr/ ISfinder] emerged from the IS''NCY'' orphan group. It is based on both Tpase and '''IR''' sequence similarities [[:File:ISAzo13.1.png|(Fig.ISAzo13)]]. Insertion generates a 3bp AT-rich DR and the ends have a consensus GGa/g. Their Tpases are highly conserved with a probable DDE motif and an [[wikipedia:Helix-turn-helix|HTH motif]] at the N-terminus which could function as a DNA binding domain. Two members encode two orfs with a possible PRF ('''P'''rogrammed '''T'''ranscriptional '''F'''rameshifting) motif of 8 or 9 '''A''' while the other members encode a unique orf which includes a triple lysine at the equivalent position (Gourbeyre, unpublished)<ref><pubmed>16381877</pubmed></ref>. | |
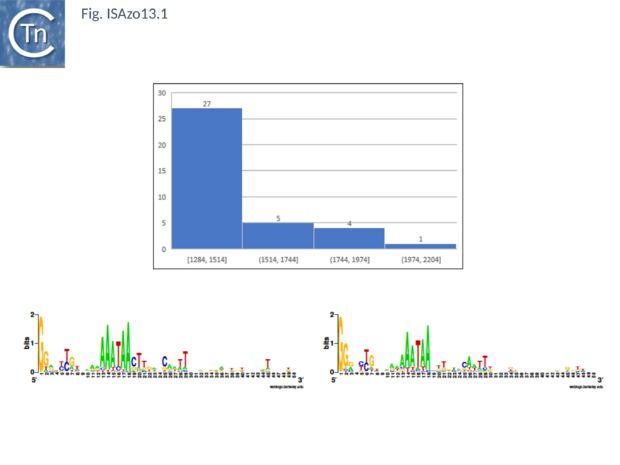
| − | [[Image:ISAzo13.1.png|thumb|center| | + | [[Image:ISAzo13.1.png|thumb|center|620x620px|'''Fig. ISAzo13.''' '''General [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAzo13 IS''Azo13''] characteristics, average length and common ends. Top:''' Distribution of IS length (base pairs) of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAzo13 IS''Azo13''] family members. The number of examples used in the sample is shown above each column. '''Bottom''': Left ('''IRL''') and right '''IRR''' inverted terminal repeats are shown in [http://weblogo.threeplusone.com/ WebLogo] format.|alt=]] |
| + | ==Bibliography== | ||
| + | {{Reflist|32em}} | ||
| + | <br /> | ||
| + | <hr> | ||
| + | {{TnPedia}} | ||
Latest revision as of 12:55, 5 December 2022
This family, represented by 37 members in ISfinder emerged from the ISNCY orphan group. It is based on both Tpase and IR sequence similarities (Fig.ISAzo13). Insertion generates a 3bp AT-rich DR and the ends have a consensus GGa/g. Their Tpases are highly conserved with a probable DDE motif and an HTH motif at the N-terminus which could function as a DNA binding domain. Two members encode two orfs with a possible PRF (Programmed Transcriptional Frameshifting) motif of 8 or 9 A while the other members encode a unique orf which includes a triple lysine at the equivalent position (Gourbeyre, unpublished)[1].

Fig. ISAzo13. General ISAzo13 characteristics, average length and common ends. Top: Distribution of IS length (base pairs) of ISAzo13 family members. The number of examples used in the sample is shown above each column. Bottom: Left (IRL) and right IRR inverted terminal repeats are shown in WebLogo format.
Bibliography