Difference between revisions of "IS Families/IS21 family"
| Line 1: | Line 1: | ||
====General==== | ====General==== | ||
| − | IS''21'' was discovered in plasmid R68 where it was subsequently observed to undergo a tandem duplication which greatly facilitated the insertion of the plasmid into the ''[[wikipedia:Pseudomonas|Pseudomonas]]'' host genome. This led to the formation of Hfr strains<ref name=":0"><pubmed>6777257</pubmed> | + | IS''21'' was discovered in plasmid R68 where it was subsequently observed to undergo a tandem duplication which greatly facilitated the insertion of the plasmid into the ''[[wikipedia:Pseudomonas|Pseudomonas]]'' host genome. This led to the formation of Hfr strains<ref name=":0"><pubmed>6777257</pubmed> |
| − | |||
| − | + | </nowiki></ref><ref><nowiki><pubmed>6269130</pubmed></nowiki></ref> and the subsequent demonstration that the ''[[wikipedia:Pseudomonas|Pseudomonas]]'' chromosome was genetically circular<ref><nowiki><pubmed>417059</pubmed></nowiki></ref> [[:File:Fig. IS21.1.png|(Fig.IS21.1)]]. | |
| + | [[Image:Fig. IS21.1.png|thumb|center|620x620px|'''Fig. IS21.1.''' '''IS''21'' family common mechanism'''. IS''21'' is thought to transpose using a '''copy-out-paste-in mechanism''' via a circular intermediate ('''left''') The original [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] identified in R66 has only low activity due to an inefficient transposase promoter. An R66 derivative was isolated carrying an [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] tandem dimer ('''right'''). This creates an active junction similar to that in the circle together with a hybrid junction promoter with a -35 in one end and a -10 promoter element in the other. This drives transposase expression from the second IS.|alt=]] | ||
| − | + | The conjugative plasmid R68 carries a single copy of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] and is inefficient in chromosome transfer <ref name=":0" />. An R68 derivative, R68.45, exhibiting a marked increase in transfer of chromosomal genes, was isolated <ref name=":0" /> and proved to carry two directly repeated copies of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] separated by 3 bp<ref name=":1"><pubmed>2540414</pubmed></nowiki></ref> [[:File:Fig. IS21.1.png|(Fig.IS21.1)]]. | |
| − | + | In addition to the distinct examples restricted to individual species, several individual ISs are themselves quite widely distributed. Isoforms of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''], several of which are incomplete, have been found in RP4 ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS8]<ref><nowiki><pubmed>6253354</pubmed></nowiki></ref>), in ''[[wikipedia:Shigella_sonnei|Shigella sonnei]]'', and various clinical isolates of ''[[wikipedia:Escherichia_coli|Escherichia coli]]'', in the EHEC plasmid p0157<ref><nowiki><pubmed>9722640</pubmed></nowiki></ref> and in the ''[[wikipedia:Yersinia_pestis|Yersinia pestis]]'' virulence plasmid pCD1 ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS640 IS''640'']<ref><nowiki><pubmed>9748454</pubmed></nowiki></ref>; [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21p'']). Another family member, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS100 IS''100''] is found in several ''[[wikipedia:Yersinia|Yersinia]]'' plasmids, including pCD1<ref><nowiki><pubmed>8833859</pubmed></nowiki></ref>, flanking a high pathogenicity island in ''[[wikipedia:Yersinia_pestis|Y. pestis]]'' (as well as multiple copies in the genome) and ''[[wikipedia:Yersinia_pseudotuberculosis|Y. pseudotuberculosis]]''<ref><nowiki><pubmed>9988474</pubmed></nowiki></ref>, and in the uropathogenic ''[[wikipedia:Escherichia_coli|E. coli]]'' J96<ref><nowiki><pubmed>8751923</pubmed></nowiki></ref>. | |
| − | |||
| − | [[ | ||
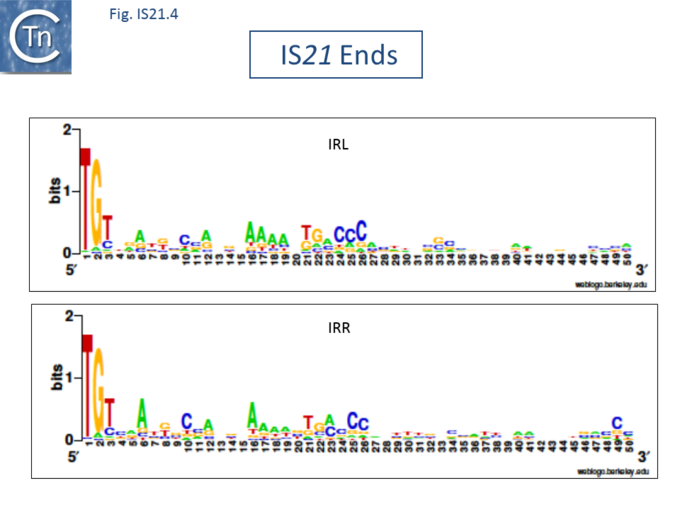
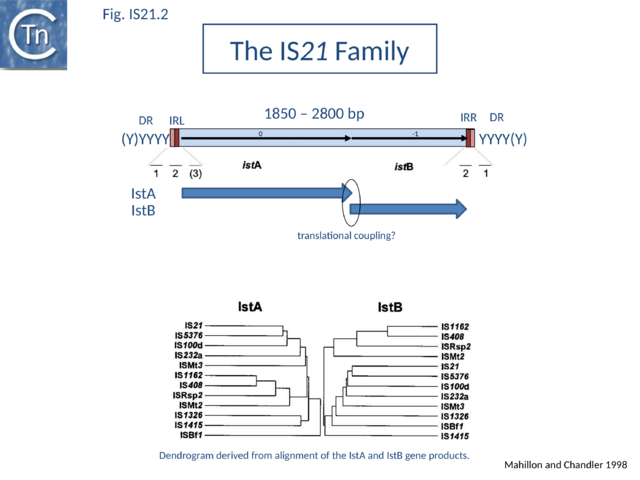
| − | + | The IS''21'' family is fairly homogeneous. Family members have lengths in the range 1850-2800bp and are therefore among the largest bacterial IS elements ([[General Information/What Is an IS?#Characteristics%20of%20insertion%20sequence%20families|TABLE Characteristics of IS families]]). They carry related terminal IRs whose lengths may vary between 11 (e.g., [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21'']) and 50 bp (e.g., [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5376 IS''5376'']) and generally terminate in the dinucleotide 5'-CA-3' ([[:File:Fig. IS21.2.png|Fig.IS21.2]]; [[:File:Fig. IS21.3.png|IS21.3]] and [[:File:Fig. IS21.4.png|.IS21.4]]). | |
| + | [[Image:Fig. IS21.2.png|thumb|center|640x640px|'''Fig. IS21.2.''' The terminal '''IRL''' and '''IRR''' are shown as solid boxes. The position of the ''istA'' and ''istB'' reading frames is also shown. The horizontal lines below show the relative positions of multiply repeated sequences IstA with a potential DDE motif, and IstB are indicated below. The possibility of translational coupling between the two reading frames is indicated.|alt=]] | ||
| + | [[Image:Fig. IS21.3.png|thumb|680x680px|'''Fig. IS21.3.''' Nucleotide sequences of the multiple terminal repeats, together with their coordinates. CS, complementary strand. L1, L2, and L3, and R1 and R2, indicate internal repeated sequences at the left and right ends, respectively.|alt=|center]]The IS''21'' family terminal '''IRs''' are complex. Several carry multiple repeated sequences at their ends which include part of the terminal '''IRs'''<ref name=":2"><pubmed>9729608</pubmed> | ||
| − | IS''21'' | + | </nowiki></ref> ([[:File:Fig. IS21.2.png|Fig.IS21.2]]) and which may represent Tpase binding sites. IS''21'' itself also carries a degenerate set of such sites ([https://renouvaud.hosted.exlibrisgroup.com/primo-explore/fulldisplay?docid=41BCU_ALMA7155645620002851&context=L&vid=41BCULIB_VU2&lang=fr_FR&search_scope=41BCULIB_ALMA_ALL&adaptor=Local%20Search%20Engine&tab=default_tab&query=lsr04,exact,R269895760 Berger B, 2000, La séquence d’insertion IS''21'': la famille de cette élément bactérien, sa spécificité d'insertion et son utilisation pour une méthode de linker insertion mutagenesis ''in vitro''], [https://www.unil.ch/index.html Université de Lausanne] undergraduate thesis) which were not initially identified. Like a number of IS families, the ends of the element terminate with the dinucleotide 5’-CA-3’ ([[:File:Fig. IS21.4.png|Fig.IS21.4]]). |
| − | + | Insertion results in a direct target repeat of 4 or, more frequently, 5 bp while two members (eg., [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS53 IS''53''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS408 IS''408'']) may generate 8 bp. | |
| − | + | IS''21'' family members encode two consecutive genes, along upstream frame designated ''istA'' the Tpase and a “'''helper'''” gene, and a shorter downstream frame, ''istB'' ([[:File:Fig. IS21.2.png|Fig.IS21.2]]) which exhibits some similarity to the [[wikipedia:DnaA|DnaA]] replication initiator protein due to the presence of an [[wikipedia:ATP-binding_motif|ATP binding motif]], and often appears in BLAST searches of complete genomes. | |
| − | + | Overall identities range from 10 to 59% for IstA and from 25 to 67% for IstB. For one member, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS53 IS''53'']<ref><nowiki><pubmed>8385786</pubmed></nowiki></ref>, a match of 45% identity of the potential ''istA'' frame with that of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1326 IS''1326''] can be obtained by the introduction of several frameshifts. An additional sequence, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS640 IS''640'']<ref><nowiki><pubmed>2824781</pubmed></nowiki></ref> from ''[[wikipedia:Shigella_sonnei|Shigella sonnei]]'', is a truncated iso-[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21'']. It carries an ''istA'' reading frame which is nearly identical to that of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] but is lacking ''istB'' and the right terminal IR. Many additional [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] derivatives are appearing in the various bacterial genome sequences. | |
| − | [[ | + | The ''istB'' frame may be located in a relative reading phase of -1 (e.g. [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5376 IS''5376'']) or +1 (e.g. [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS232 IS''232''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1326 IS''1326'']) compared to ''istA''. It can be slightly separated from ''istA'' (17 bp, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS408 IS''408''] - for which only a partial sequence lacking the right end is available) or overlap for one ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21'']) or several base pairs ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS232 IS''232''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5376 IS''5376''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1326 IS''1326'']), and is generally preceded by a potential ribosome binding site. In many members, the termination codon of ''istA'' and the initiation codon of ''istB'' overlap suggesting that IstB is produced by translational coupling ([[:File:Fig. IS21.2.png|Fig.IS21.2]]). |
| − | < | + | The ''istA'' reading frame carries a motif related to the widespread [[wikipedia:Integrase|integrase]] DDE motif but lacking the conserved K/R residue and a potential helix-turn-helix motif. Although there is no general conservation of the K/R residue expected seven residues downstream from the final '''E''' , there is good conservation of a basic amino acid at either position 7 or 8 ([https://renouvaud.hosted.exlibrisgroup.com/primo-explore/fulldisplay?docid=41BCU_ALMA7155645620002851&context=L&vid=41BCULIB_VU2&lang=fr_FR&search_scope=41BCULIB_ALMA_ALL&adaptor=Local%20Search%20Engine&tab=default_tab&query=lsr04,exact,R269895760 Berger B, 2000, La séquence d’insertion IS21: la famille de cette élément bactérien, sa spécificité d'insertion et son utilisation pour une méthode de linker insertion mutagenesis ''in vitro''], Université de Lausanne undergraduate thesis). The ''istB'' frame carries a relatively well conserved potential NTP binding domain<ref><nowiki><pubmed>1533715</pubmed></nowiki></ref>. Sequence alignments of IstA and of IstB gives very similar relationships <ref name=":2" />. |
| + | |||
| + | [[Image:Fig. IS21.4.png|thumb|center|680x680px|'''Fig. IS21.4.''' '''[http://weblogo.threeplusone.com/ Weblogo] showing the common IS''21'' ends.''' Left ('''IRL''') and right '''IRR''' inverted terminal repeats are shown in [http://weblogo.threeplusone.com/ WebLogo] format.|alt=]] | ||
=====IS''21''===== | =====IS''21''===== | ||
| − | IS''21'' has been studied in some detail. The tandem duplication observed in the original R68.45<ref name=":1" /> can be envisaged to occur by various routes. It is presumably favoured by the marked tendency of IS''21'' to insert in, or close to, an IS''21'' end<ref name=":3"><pubmed>3034717</pubmed> | + | IS''21'' has been studied in some detail. The tandem duplication observed in the original R68.45<ref name=":1" /> can be envisaged to occur by various routes. It is presumably favoured by the marked tendency of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] to insert in, or close to, an [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] end<ref name=":3"><pubmed>3034717</pubmed> |
| + | |||
| + | </nowiki></ref>. It may also occur by unequal crossing over during replication of the donor plasmid or, as suggested for [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS30 IS''30''], whose transposition mechanism shows some similarity with that of IS''21'' (see "[[IS Families/IS30 family|IS''30'' family]]"), by dimerization of the donor plasmid and deletion of one intervening plasmid sequence located between the two IS copies ([[:File:Fig. IS21.1.png|Fig.IS21.1]]). This type of mechanism has been demonstrated experimentally both for [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS30 IS''30'']<ref><nowiki><pubmed>10540284</pubmed></nowiki></ref> and for [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS911 IS''911'']<ref><nowiki><pubmed>10760133</pubmed></nowiki></ref>; [[IS Families/IS3 family|IS''3'' family]]). The tandemly repeated copies of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] promote insertion of the entire plasmid in a transposition event involving the abutted terminal IRs. Integration results in loss of the 3 bp located between the two ends in the parental R68.45 plasmid. | ||
The resulting replicon fusion thus has the appearance of a true cointegrate (i.e. with a single directly repeated copy of the IS at each junction) although transposition does not involve duplication of the insertion sequence during the insertion event. | The resulting replicon fusion thus has the appearance of a true cointegrate (i.e. with a single directly repeated copy of the IS at each junction) although transposition does not involve duplication of the insertion sequence during the insertion event. | ||
| − | Both IstA and IstB are required for integration<ref name=":4"><pubmed>2174355</pubmed> | + | Both IstA and IstB are required for integration<ref name=":4"><pubmed>2174355</pubmed></nowiki></ref> and production of a second IstA derivative (from an alternative translational initiation codon eight codons downstream of that of IstA itself) greatly increases the efficiency of replicon fusions <ref name=":4" /> for reasons that are at present unknown Integration is optimal when the distance separating the two ends is 4 bp. It is efficient with a 2 or 3 base pair separation but inefficient with smaller or larger intervening sequences<ref name=":5"><pubmed>9737923</pubmed></nowiki></ref>. Like retroviruses, and several other transposable elements, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] terminates with a 5'-CA-3'. Mutation of the terminal A abolishes replicon fusion (Haas et al., 1996, '''Insertion sequence IS''21'': Related insertion sequence elements, transpositional mechanisms, and application to linker insertion mutagenesis''', in [https://www.amazon.com.au/Molecular-Biology-Pseudomonads-T-Nakazawa/dp/1555811043 Nakazawa T, Molecular biology of ''Pseudomonads'']).[[Image:Fig. IS21.7.png|thumb|720x720px|'''Fig. IS21.7.''' '''The model of IS''21'' transposition cycle.''' The DDE transposase, IstA is shown to carry out all steps in the copy-out-paste-in transposition pathway on its own to a step before target capture. At this point, the ATPase IstB intervenes together with IstA. Based on Arias-Palomo and Berger, 2015. |alt=|center]] |
| − | The duplication of IS''21'' in R68.45 not only creates an efficient integrative donor structure by providing two closely spaced and correctly oriented IRs, but generates a strong promoter (as measured with a ''[https://www.wikigenes.org/e/gene/e/945358.html galK]'' [[wikipedia:Reporter_gene|reporter gene]]) in which a -35 box located in the IRR of the upstream element is combined with a -10 box located in the abutted IRL of the downstream copy. This directs high levels of production of the IS''21'' transposition proteins <ref name=":1" /> in a similar manner to the junction promoter created by circularization of IS''911'' and IS''2''. The insertion of IS''21'' downstream of strong promoter results in the efficient expression of transposition proteins. Thus IS''21'' appears not to have adopted a mechanism which "protects" it against high levels of impinging transcription. It is presumably the necessity for a transposition substrate carrying two abutted ends and the low frequency of formation of such molecules which acts as a control on transposition activity. More recently, a low level of circular IS''21'' copies has been detected in preparations of IS''21''-carrying plasmid DNA<ref name=":6"><pubmed>11315188</pubmed> | + | The duplication of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] in R68.45 not only creates an efficient integrative donor structure by providing two closely spaced and correctly oriented IRs, but generates a strong promoter (as measured with a ''[https://www.wikigenes.org/e/gene/e/945358.html galK]'' [[wikipedia:Reporter_gene|reporter gene]]) in which a -35 box located in the IRR of the upstream element is combined with a -10 box located in the abutted IRL of the downstream copy. This directs high levels of production of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] transposition proteins <ref name=":1" /> in a similar manner to the junction promoter created by circularization of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS911 IS''911''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS2 IS''2'']. The insertion of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] downstream of strong promoter results in the efficient expression of transposition proteins. Thus [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] appears not to have adopted a mechanism which "protects" it against high levels of impinging transcription. It is presumably the necessity for a transposition substrate carrying two abutted ends and the low frequency of formation of such molecules which acts as a control on transposition activity. More recently, a low level of circular [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] copies has been detected in preparations of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21'']-carrying plasmid DNA<ref name=":6"><pubmed>11315188</pubmed></nowiki></ref> ([https://pubmed.ncbi.nlm.nih.gov/9737923 S. Schmid and D. Haas], pers.comm.), raising the possibility that [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''], like at least some of the [[IS Families/IS3 family|IS''3'' family members]], may also transpose via a circular intermediate. |
| − | It has been demonstrated ''in vitro'', using ''[[wikipedia:Escherichia_coli|E. coli]]'' cell extracts enriched for IstA, that this protein is responsible for the 3' end cleavage of IS''21<ref name=":4" />''. A substrate carrying two abutted IS''21'' ends separated by 3 bp is cleaved specifically at both 3' ends in an IstA-dependent way [see <ref name=":6" />]. | + | It has been demonstrated ''in vitro'', using ''[[wikipedia:Escherichia_coli|E. coli]]'' cell extracts enriched for IstA, that this protein is responsible for the 3' end cleavage of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21'']''<ref name=":4" />''. A substrate carrying two abutted [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] ends separated by 3 bp is cleaved specifically at both 3' ends in an IstA-dependent way [see <ref name=":6" />]. |
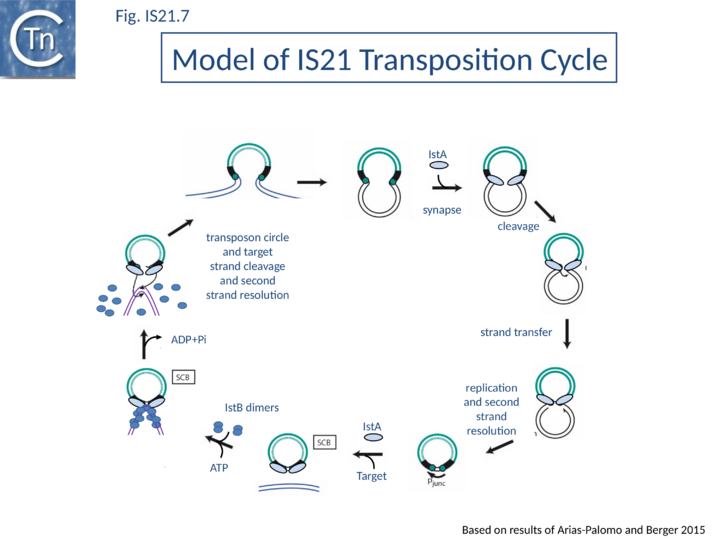
| − | Current data are consistent with the notion that IS''21'' family members transpose using a two-step mechanism ([[:File:Fig. IS21.7.png|Fig.IS21.7]]) by the formation of a reactive junction, similar to those formed in the copy-paste mechanism of IS''3'' and other families, in which two abutted IS''21'' ends are separated by several nucleotides. This is consistent with the marked tendency of IS''21'' to insert in, or close to an IS''21'' end. The reactive junction is subsequently integrated into the target DNA to give a DR of 4 to 8 bp. Integration is optimal when the distance separating the two ends in the junction is 4 bp. It is efficient with a 2- or 3-bp separation but inefficient with smaller or larger intervening sequences. IstA carries the DDE motif and is the Tpase. IstB is necessary for the target DNA capture and accurate insertion by IstA donor insertion by its cognate transposase <ref name=":4" />. | + | Current data are consistent with the notion that [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] family members transpose using a two-step mechanism ([[:File:Fig. IS21.7.png|Fig.IS21.7]]) by the formation of a reactive junction, similar to those formed in the copy-paste mechanism of [[IS Families/IS3 family|IS''3'' and other families]], in which two abutted [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] ends are separated by several nucleotides. This is consistent with the marked tendency of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] to insert in, or close to an [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] end. The reactive junction is subsequently integrated into the target DNA to give a DR of 4 to 8 bp. Integration is optimal when the distance separating the two ends in the junction is 4 bp. It is efficient with a 2- or 3-bp separation but inefficient with smaller or larger intervening sequences. IstA carries the DDE motif and is the Tpase. IstB is necessary for the target DNA capture and accurate insertion by IstA donor insertion by its cognate transposase <ref name=":4" />. |
Both IstA and IstB are required for efficient transposition while a product of alternative natural translation initiation within the Tpase gene (which removes the eight N-terminal amino acids) may facilitate integration to generate “cointegrates” and has been called a “cointegrase”<ref name=":5" />. | Both IstA and IstB are required for efficient transposition while a product of alternative natural translation initiation within the Tpase gene (which removes the eight N-terminal amino acids) may facilitate integration to generate “cointegrates” and has been called a “cointegrase”<ref name=":5" />. | ||
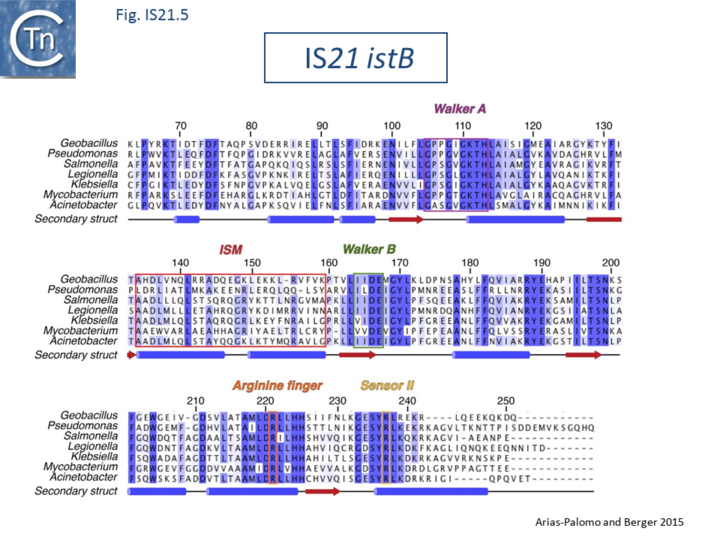
| − | Biochemical and structural data are now available concerning the role of IstB in IS''21'' transposition<ref name=":7"><pubmed>26276634</pubmed> | + | Biochemical and structural data are now available concerning the role of IstB in [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] transposition<ref name=":7"><pubmed>26276634</pubmed></nowiki></ref>. Several transposable elements use regulatory AAA+ ATPases for function. For [[wikipedia:Bacteriophage_Mu|bacteriophage Mu]] (MuB) and [[Transposons families/Tn7 family|Tn''7'']] (TnsC), these proteins are involved in the choice of target sites<ref><nowiki><pubmed>26104363</pubmed></nowiki></ref><ref><nowiki><pubmed>26104374</pubmed></nowiki></ref>. [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''], also carries a conserved potential NTP binding domain ([[:File:Fig. IS21.5.png|Fig.IS21.5]]) which shares sequence similarity with the [[wikipedia:DnaA|DnaA]] protein and because of this, often interferes with automatic bacterial genome annotation pipelines. |
| − | [[Image:Fig. IS21.5.png|thumb|center| | + | [[Image:Fig. IS21.5.png|thumb|center|720x720px|'''Fig. IS21.5.''' Alignment of IS''21'' ATPase IstB. Adapted from Arias-Palomo and Berger, 2015. IstB CDS were identified in the bacteria listed on the left. Walker A and B boxes are indicated in addition to an arginine finger, and a C-terminal sensor domain common in AAA+ proteins. IstB also includes another characteristic of AAA+ proteins, an initiator/loader specific motif (ISM) Dueber et al., 2007; Iyer et al., 2004). Details can be found in Arias-Palomo and Berger 2015.|alt=]] |
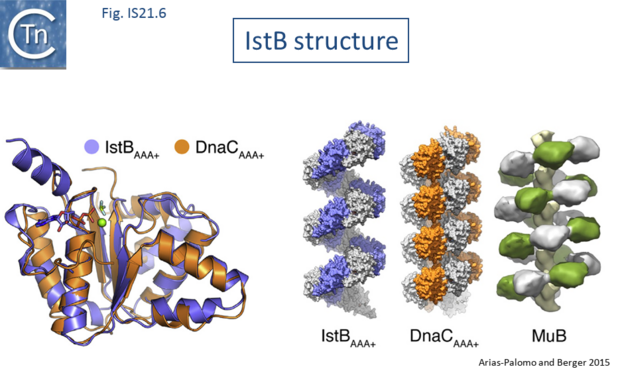
The [[wikipedia:X-ray_crystallography|X-ray crystallographic]] structure of IstB with its N-terminal domain removed including a bound inactive ADP-BeF3 ligand superposed well onto the AAA+ domain of DnaC ([[:File:Fig. IS21.6.png|Fig.IS21.6]]). X-ray crystallography and [[wikipedia:Cryogenic_electron_microscopy|cryo-electron microscopy]] showed that the full length IstB protein self-assembles into a helical decamer (a pentamer of dimers; a so-called clamshell-shape) forming a right-handed filament very similar to that of [[wikipedia:DnaC|DnaC]] and MuB<ref><nowiki><pubmed>23776210</pubmed></nowiki></ref> ([[:File:Fig. IS21.6.png|Fig.IS21.6]]). Multimerization is dependent on the ATP-co-ordinating residues. This filament was capable of grasping DNA between two pentamers generating a 180° bend and also to introduce positive supercoils in an ATP dependent manner presumably by wrapping the DNA. Binding appears to show no sequence-specificity. | The [[wikipedia:X-ray_crystallography|X-ray crystallographic]] structure of IstB with its N-terminal domain removed including a bound inactive ADP-BeF3 ligand superposed well onto the AAA+ domain of DnaC ([[:File:Fig. IS21.6.png|Fig.IS21.6]]). X-ray crystallography and [[wikipedia:Cryogenic_electron_microscopy|cryo-electron microscopy]] showed that the full length IstB protein self-assembles into a helical decamer (a pentamer of dimers; a so-called clamshell-shape) forming a right-handed filament very similar to that of [[wikipedia:DnaC|DnaC]] and MuB<ref><nowiki><pubmed>23776210</pubmed></nowiki></ref> ([[:File:Fig. IS21.6.png|Fig.IS21.6]]). Multimerization is dependent on the ATP-co-ordinating residues. This filament was capable of grasping DNA between two pentamers generating a 180° bend and also to introduce positive supercoils in an ATP dependent manner presumably by wrapping the DNA. Binding appears to show no sequence-specificity. | ||
| + | |||
| + | <br /> | ||
[[Image:Fig. IS21.6.png|thumb|center|620x620px|'''Fig. IS21.6.''' '''The IstB structure.''' Taken from Arias-Palomo and Berger, 2015. Left: Superposition of InsB and the AAA+ replication initiator protein DnaC. Right: right-handed filament formed by IstB compared with other ATpases, DnaC and the MuB ATPase involved in target capture by the transposable bacteriophage Mu. Details can be found in Arias-Palomo and Berger 2015.|alt=]] | [[Image:Fig. IS21.6.png|thumb|center|620x620px|'''Fig. IS21.6.''' '''The IstB structure.''' Taken from Arias-Palomo and Berger, 2015. Left: Superposition of InsB and the AAA+ replication initiator protein DnaC. Right: right-handed filament formed by IstB compared with other ATpases, DnaC and the MuB ATPase involved in target capture by the transposable bacteriophage Mu. Details can be found in Arias-Palomo and Berger 2015.|alt=]] | ||
The transposase, IstA, was also shown to recognize the IstB DNA complex and to promote disassembly by stimulating ATP hydrolysis. | The transposase, IstA, was also shown to recognize the IstB DNA complex and to promote disassembly by stimulating ATP hydrolysis. | ||
| − | A model has been proposed in which IstB binding and subsequent DNA bending primes a selected insertion site for efficient transposition: an IstB decamer first associates with target DNA, and the complex then recruits IstA in the form of a transpososome or paired-end complex (PEC) including a pair of inverted IS''21'' repeats to generate a synaptic complex. Although it has been proposed that the PEC has precleared ends, this is not an essential requirement. The order of strand cleavage and the multiple stages of transpososome assembly must yet be defined. The inability of DNA to promote ATP turnover by IstB would impart sufficient stability and allow time for IstA recognition. IstA-stimulated ATP hydrolysis would lead to IstB dissociation to a clearing of the DNA, allowing the IstA access to the target DNA. In this model, IstB might be considered an AAA+-type co-chaperone<ref name=":7" /> ([[:File:Fig. IS21.7.png|Fig.IS21.7]]). | + | A model has been proposed in which IstB binding and subsequent DNA bending primes a selected insertion site for efficient transposition: an IstB decamer first associates with target DNA, and the complex then recruits IstA in the form of a transpososome or paired-end complex (PEC) including a pair of inverted [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] repeats to generate a synaptic complex. Although it has been proposed that the PEC has precleared ends, this is not an essential requirement. The order of strand cleavage and the multiple stages of transpososome assembly must yet be defined. The inability of DNA to promote ATP turnover by IstB would impart sufficient stability and allow time for IstA recognition. IstA-stimulated ATP hydrolysis would lead to IstB dissociation to a clearing of the DNA, allowing the IstA access to the target DNA. In this model, IstB might be considered an AAA+-type co-chaperone<ref name=":7" /> ([[:File:Fig. IS21.7.png|Fig.IS21.7]]). |
| − | IS''21'' has been reported to show a preference for insertion close to the end of a second copy of the element located in the target plasmid<ref name=":3" />. This is a characteristic of many IS families whose members transpose via a circular dsDNA intermediate and presumably reflects their ability to form a synaptic complex between an IR carried by the intact IS and a second IR in the target molecule (see [[:File:Fig. IS3.11.png|Fig.IS3.11]]). | + | [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS21 IS''21''] has been reported to show a preference for insertion close to the end of a second copy of the element located in the target plasmid<ref name=":3" />. This is a characteristic of many IS families whose members transpose via a circular dsDNA intermediate and presumably reflects their ability to form a synaptic complex between an IR carried by the intact IS and a second '''IR''' in the target molecule (see [[:File:Fig. IS3.11.png|Fig.IS3.11]]). |
<br /> | <br /> | ||
==Bibliography== | ==Bibliography== | ||
<references /> | <references /> | ||
Revision as of 17:54, 11 August 2021
General
IS21 was discovered in plasmid R68 where it was subsequently observed to undergo a tandem duplication which greatly facilitated the insertion of the plasmid into the Pseudomonas host genome. This led to the formation of Hfr strains[1][2] and the subsequent demonstration that the Pseudomonas chromosome was genetically circular[3] (Fig.IS21.1).
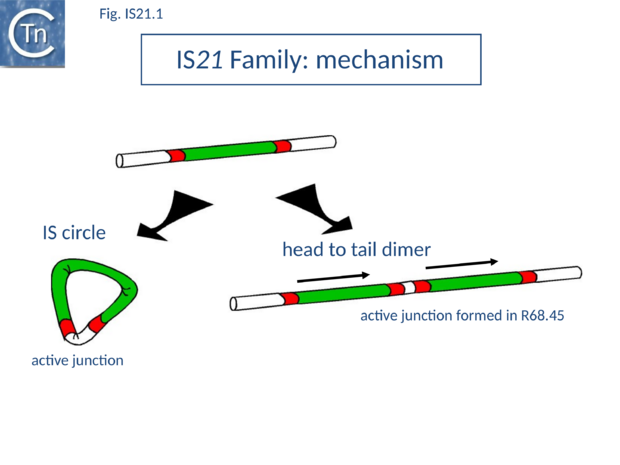
The conjugative plasmid R68 carries a single copy of IS21 and is inefficient in chromosome transfer [1]. An R68 derivative, R68.45, exhibiting a marked increase in transfer of chromosomal genes, was isolated [1] and proved to carry two directly repeated copies of IS21 separated by 3 bp[4] (Fig.IS21.1).
In addition to the distinct examples restricted to individual species, several individual ISs are themselves quite widely distributed. Isoforms of IS21, several of which are incomplete, have been found in RP4 (IS8[5]), in Shigella sonnei, and various clinical isolates of Escherichia coli, in the EHEC plasmid p0157[6] and in the Yersinia pestis virulence plasmid pCD1 (IS640[7]; IS21p). Another family member, IS100 is found in several Yersinia plasmids, including pCD1[8], flanking a high pathogenicity island in Y. pestis (as well as multiple copies in the genome) and Y. pseudotuberculosis[9], and in the uropathogenic E. coli J96[10].
The IS21 family is fairly homogeneous. Family members have lengths in the range 1850-2800bp and are therefore among the largest bacterial IS elements (TABLE Characteristics of IS families). They carry related terminal IRs whose lengths may vary between 11 (e.g., IS21) and 50 bp (e.g., IS5376) and generally terminate in the dinucleotide 5'-CA-3' (Fig.IS21.2; IS21.3 and .IS21.4).
The IS21 family terminal IRs are complex. Several carry multiple repeated sequences at their ends which include part of the terminal IRs[11] (Fig.IS21.2) and which may represent Tpase binding sites. IS21 itself also carries a degenerate set of such sites (Berger B, 2000, La séquence d’insertion IS21: la famille de cette élément bactérien, sa spécificité d'insertion et son utilisation pour une méthode de linker insertion mutagenesis in vitro, Université de Lausanne undergraduate thesis) which were not initially identified. Like a number of IS families, the ends of the element terminate with the dinucleotide 5’-CA-3’ (Fig.IS21.4).
Insertion results in a direct target repeat of 4 or, more frequently, 5 bp while two members (eg., IS53 and IS408) may generate 8 bp.
IS21 family members encode two consecutive genes, along upstream frame designated istA the Tpase and a “helper” gene, and a shorter downstream frame, istB (Fig.IS21.2) which exhibits some similarity to the DnaA replication initiator protein due to the presence of an ATP binding motif, and often appears in BLAST searches of complete genomes.
Overall identities range from 10 to 59% for IstA and from 25 to 67% for IstB. For one member, IS53[12], a match of 45% identity of the potential istA frame with that of IS1326 can be obtained by the introduction of several frameshifts. An additional sequence, IS640[13] from Shigella sonnei, is a truncated iso-IS21. It carries an istA reading frame which is nearly identical to that of IS21 but is lacking istB and the right terminal IR. Many additional IS21 derivatives are appearing in the various bacterial genome sequences.
The istB frame may be located in a relative reading phase of -1 (e.g. IS21, IS5376) or +1 (e.g. IS232, IS1326) compared to istA. It can be slightly separated from istA (17 bp, IS408 - for which only a partial sequence lacking the right end is available) or overlap for one (IS21) or several base pairs (IS232, IS5376, IS1326), and is generally preceded by a potential ribosome binding site. In many members, the termination codon of istA and the initiation codon of istB overlap suggesting that IstB is produced by translational coupling (Fig.IS21.2).
The istA reading frame carries a motif related to the widespread integrase DDE motif but lacking the conserved K/R residue and a potential helix-turn-helix motif. Although there is no general conservation of the K/R residue expected seven residues downstream from the final E , there is good conservation of a basic amino acid at either position 7 or 8 (Berger B, 2000, La séquence d’insertion IS21: la famille de cette élément bactérien, sa spécificité d'insertion et son utilisation pour une méthode de linker insertion mutagenesis in vitro, Université de Lausanne undergraduate thesis). The istB frame carries a relatively well conserved potential NTP binding domain[14]. Sequence alignments of IstA and of IstB gives very similar relationships [11].
IS21
IS21 has been studied in some detail. The tandem duplication observed in the original R68.45[4] can be envisaged to occur by various routes. It is presumably favoured by the marked tendency of IS21 to insert in, or close to, an IS21 end[15]. It may also occur by unequal crossing over during replication of the donor plasmid or, as suggested for IS30, whose transposition mechanism shows some similarity with that of IS21 (see "IS30 family"), by dimerization of the donor plasmid and deletion of one intervening plasmid sequence located between the two IS copies (Fig.IS21.1). This type of mechanism has been demonstrated experimentally both for IS30[16] and for IS911[17]; IS3 family). The tandemly repeated copies of IS21 promote insertion of the entire plasmid in a transposition event involving the abutted terminal IRs. Integration results in loss of the 3 bp located between the two ends in the parental R68.45 plasmid.
The resulting replicon fusion thus has the appearance of a true cointegrate (i.e. with a single directly repeated copy of the IS at each junction) although transposition does not involve duplication of the insertion sequence during the insertion event.
Both IstA and IstB are required for integration[18] and production of a second IstA derivative (from an alternative translational initiation codon eight codons downstream of that of IstA itself) greatly increases the efficiency of replicon fusions [18] for reasons that are at present unknown Integration is optimal when the distance separating the two ends is 4 bp. It is efficient with a 2 or 3 base pair separation but inefficient with smaller or larger intervening sequences[19]. Like retroviruses, and several other transposable elements, IS21 terminates with a 5'-CA-3'. Mutation of the terminal A abolishes replicon fusion (Haas et al., 1996, Insertion sequence IS21: Related insertion sequence elements, transpositional mechanisms, and application to linker insertion mutagenesis, in Nakazawa T, Molecular biology of Pseudomonads).
The duplication of IS21 in R68.45 not only creates an efficient integrative donor structure by providing two closely spaced and correctly oriented IRs, but generates a strong promoter (as measured with a galK reporter gene) in which a -35 box located in the IRR of the upstream element is combined with a -10 box located in the abutted IRL of the downstream copy. This directs high levels of production of the IS21 transposition proteins [4] in a similar manner to the junction promoter created by circularization of IS911 and IS2. The insertion of IS21 downstream of strong promoter results in the efficient expression of transposition proteins. Thus IS21 appears not to have adopted a mechanism which "protects" it against high levels of impinging transcription. It is presumably the necessity for a transposition substrate carrying two abutted ends and the low frequency of formation of such molecules which acts as a control on transposition activity. More recently, a low level of circular IS21 copies has been detected in preparations of IS21-carrying plasmid DNA[20] (S. Schmid and D. Haas, pers.comm.), raising the possibility that IS21, like at least some of the IS3 family members, may also transpose via a circular intermediate.
It has been demonstrated in vitro, using E. coli cell extracts enriched for IstA, that this protein is responsible for the 3' end cleavage of IS21[18]. A substrate carrying two abutted IS21 ends separated by 3 bp is cleaved specifically at both 3' ends in an IstA-dependent way [see [20]].
Current data are consistent with the notion that IS21 family members transpose using a two-step mechanism (Fig.IS21.7) by the formation of a reactive junction, similar to those formed in the copy-paste mechanism of IS3 and other families, in which two abutted IS21 ends are separated by several nucleotides. This is consistent with the marked tendency of IS21 to insert in, or close to an IS21 end. The reactive junction is subsequently integrated into the target DNA to give a DR of 4 to 8 bp. Integration is optimal when the distance separating the two ends in the junction is 4 bp. It is efficient with a 2- or 3-bp separation but inefficient with smaller or larger intervening sequences. IstA carries the DDE motif and is the Tpase. IstB is necessary for the target DNA capture and accurate insertion by IstA donor insertion by its cognate transposase [18].
Both IstA and IstB are required for efficient transposition while a product of alternative natural translation initiation within the Tpase gene (which removes the eight N-terminal amino acids) may facilitate integration to generate “cointegrates” and has been called a “cointegrase”[19].
Biochemical and structural data are now available concerning the role of IstB in IS21 transposition[21]. Several transposable elements use regulatory AAA+ ATPases for function. For bacteriophage Mu (MuB) and Tn7 (TnsC), these proteins are involved in the choice of target sites[22][23]. IS21, also carries a conserved potential NTP binding domain (Fig.IS21.5) which shares sequence similarity with the DnaA protein and because of this, often interferes with automatic bacterial genome annotation pipelines.
The X-ray crystallographic structure of IstB with its N-terminal domain removed including a bound inactive ADP-BeF3 ligand superposed well onto the AAA+ domain of DnaC (Fig.IS21.6). X-ray crystallography and cryo-electron microscopy showed that the full length IstB protein self-assembles into a helical decamer (a pentamer of dimers; a so-called clamshell-shape) forming a right-handed filament very similar to that of DnaC and MuB[24] (Fig.IS21.6). Multimerization is dependent on the ATP-co-ordinating residues. This filament was capable of grasping DNA between two pentamers generating a 180° bend and also to introduce positive supercoils in an ATP dependent manner presumably by wrapping the DNA. Binding appears to show no sequence-specificity.
The transposase, IstA, was also shown to recognize the IstB DNA complex and to promote disassembly by stimulating ATP hydrolysis.
A model has been proposed in which IstB binding and subsequent DNA bending primes a selected insertion site for efficient transposition: an IstB decamer first associates with target DNA, and the complex then recruits IstA in the form of a transpososome or paired-end complex (PEC) including a pair of inverted IS21 repeats to generate a synaptic complex. Although it has been proposed that the PEC has precleared ends, this is not an essential requirement. The order of strand cleavage and the multiple stages of transpososome assembly must yet be defined. The inability of DNA to promote ATP turnover by IstB would impart sufficient stability and allow time for IstA recognition. IstA-stimulated ATP hydrolysis would lead to IstB dissociation to a clearing of the DNA, allowing the IstA access to the target DNA. In this model, IstB might be considered an AAA+-type co-chaperone[21] (Fig.IS21.7).
IS21 has been reported to show a preference for insertion close to the end of a second copy of the element located in the target plasmid[15]. This is a characteristic of many IS families whose members transpose via a circular dsDNA intermediate and presumably reflects their ability to form a synaptic complex between an IR carried by the intact IS and a second IR in the target molecule (see Fig.IS3.11).
Bibliography
- ↑ 1.0 1.1 1.2 Riess G, Holloway BW, Pühler A . R68.45, a plasmid with chromosome mobilizing ability (Cma) carries a tandem duplication. - Genet Res: 1980 Aug, 36(1);99-109 [PubMed:6777257] [DOI] </nowiki>
- ↑ <pubmed>6269130</pubmed>
- ↑ <pubmed>417059</pubmed>
- ↑ 4.0 4.1 4.2 Reimmann C, Moore R, Little S, Savioz A, Willetts NS, Haas D . Genetic structure, function and regulation of the transposable element IS21. - Mol Gen Genet: 1989 Feb, 215(3);416-24 [PubMed:2540414] [DOI] </nowiki>
- ↑ <pubmed>6253354</pubmed>
- ↑ <pubmed>9722640</pubmed>
- ↑ <pubmed>9748454</pubmed>
- ↑ <pubmed>8833859</pubmed>
- ↑ <pubmed>9988474</pubmed>
- ↑ <pubmed>8751923</pubmed>
- ↑ 11.0 11.1 Mahillon J, Chandler M . Insertion sequences. - Microbiol Mol Biol Rev: 1998 Sep, 62(3);725-74 [PubMed:9729608] [DOI] </nowiki>
- ↑ <pubmed>8385786</pubmed>
- ↑ <pubmed>2824781</pubmed>
- ↑ <pubmed>1533715</pubmed>
- ↑ 15.0 15.1 </nowiki>
- ↑ <pubmed>10540284</pubmed>
- ↑ <pubmed>10760133</pubmed>
- ↑ 18.0 18.1 18.2 18.3 Reimmann C, Haas D . The istA gene of insertion sequence IS21 is essential for cleavage at the inner 3' ends of tandemly repeated IS21 elements in vitro. - EMBO J: 1990 Dec, 9(12);4055-63 [PubMed:2174355] [DOI] </nowiki>
- ↑ 19.0 19.1 Schmid S, Seitz T, Haas D . Cointegrase, a naturally occurring, truncated form of IS21 transposase, catalyzes replicon fusion rather than simple insertion of IS21. - J Mol Biol: 1998 Sep 25, 282(3);571-83 [PubMed:9737923] [DOI] </nowiki>
- ↑ 20.0 20.1 </nowiki>
- ↑ 21.0 21.1 Arias-Palomo E, Berger JM . An Atypical AAA+ ATPase Assembly Controls Efficient Transposition through DNA Remodeling and Transposase Recruitment. - Cell: 2015 Aug 13, 162(4);860-71 [PubMed:26276634] [DOI] </nowiki>
- ↑ <pubmed>26104363</pubmed>
- ↑ <pubmed>26104374</pubmed>
- ↑ <pubmed>23776210</pubmed>