Difference between revisions of "IS Families/IS982 family"
| Line 1: | Line 1: | ||
====General==== | ====General==== | ||
| − | '''<big>T</big>'''he IS''982'' family has over 70 entries in [https://isfinder.biotoul.fr/ ISfinder] from over 40 bacterial and archaeal species. In the case of IS''Lpl4'' from ''[[wikipedia:Lactobacillus_plantarum|Lactobacillus plantarum]]'' identical copies have been detected in ''[[wikipedia:Leuconostoc_mesenteroides|Leuconostoc mesenteroides]]'', ''[[wikipedia:Oenococcus_oeni|Oenococcus oeni]]'', and ''[[wikipedia:Lactobacillus_sakei|Lactobacillus sakei]]'' indicating horizontal gene transfer. Historically, a single IS''982'' family member, IS''Pfu3'', was first identified in the archaea from the archaea ''[[wikipedia:Pyrococcus_furiosus|Pyrococcus furiosus]]'' and this was later joined by IS''Tli1'' from ''[[wikipedia:Thermococcus_litoralis|Thermococcus litoralis]] (both mrmbero of the'' order [[wikipedia:Thermococcales|Thermococcales]]). | + | '''<big>T</big>'''he IS''982'' family has over 70 entries in [https://isfinder.biotoul.fr/ ISfinder] from over 40 bacterial and archaeal species. In the case of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISLpl4 IS''Lpl4''] from ''[[wikipedia:Lactobacillus_plantarum|Lactobacillus plantarum]]'' identical copies have been detected in ''[[wikipedia:Leuconostoc_mesenteroides|Leuconostoc mesenteroides]]'', ''[[wikipedia:Oenococcus_oeni|Oenococcus oeni]]'', and ''[[wikipedia:Lactobacillus_sakei|Lactobacillus sakei]]'' indicating horizontal gene transfer. Historically, a single [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS982 IS''982''] family member, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPfu3 IS''Pfu3''], was first identified in the archaea from the archaea ''[[wikipedia:Pyrococcus_furiosus|Pyrococcus furiosus]]'' and this was later joined by [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTli1 IS''Tli1''] from ''[[wikipedia:Thermococcus_litoralis|Thermococcus litoralis]] (both mrmbero of the'' order [[wikipedia:Thermococcales|Thermococcales]]). |
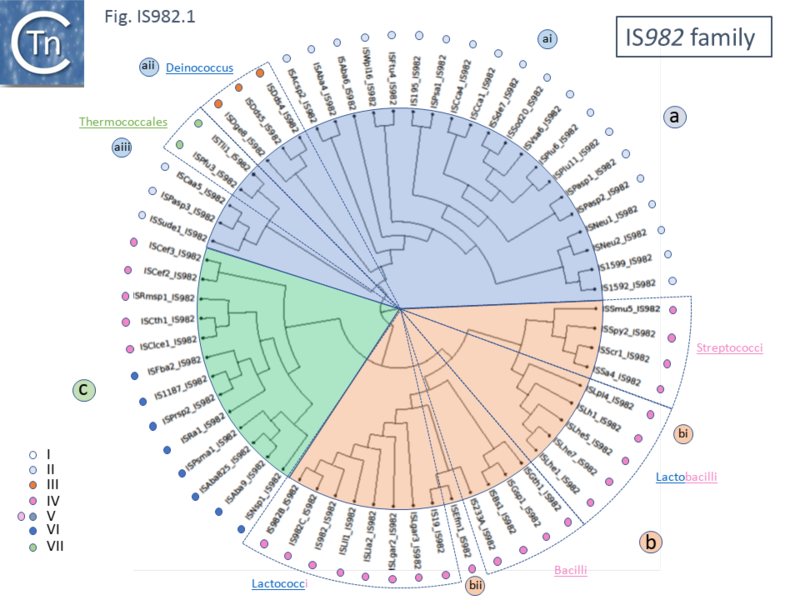
| − | A phylogenetic tree of the transposase of family members ([[:File:IS982.1.png|Fig. IS982.1]]) shows three deeply branching clades ('''a''', '''b''' and '''c''') which broadly correspond to those identified by Fayad et al. <ref name=":0"><pubmed>PMC7335449</pubmed> | + | A phylogenetic tree of the transposase of family members ([[:File:IS982.1.png|Fig. IS982.1]]) shows three deeply branching clades ('''a''', '''b''' and '''c''') which broadly correspond to those identified by Fayad et al. <ref name=":0"><pubmed>PMC7335449</pubmed> |
| + | |||
| + | </nowiki></ref>. They can be further subdivided into several subclades ('''ai'''; '''aii''' and '''aii'''; '''bi''', which includes [[wikipedia:Streptococcus|streptococci]] and [[wikipedia:Lactococcus|lactococci]]; and '''bii''' which contain bacilli and [[wikipedia:Lactobacillus|lactobacilli]]) <br /> | ||
[[File:IS982.1.png|alt=|center|thumb|800x800px|'''Fig. IS982.1.''' A dendrogram of 63 IS''982'' family members. The figure shows three major clades. The surrounding colored circles and the insert indicate the clades identified by Fayad et al. <ref name=":0" />]] | [[File:IS982.1.png|alt=|center|thumb|800x800px|'''Fig. IS982.1.''' A dendrogram of 63 IS''982'' family members. The figure shows three major clades. The surrounding colored circles and the insert indicate the clades identified by Fayad et al. <ref name=":0" />]] | ||
| − | A recent extensive [https://blast.ncbi.nlm.nih.gov/Blast.cgi BLAST] search in the non-redundant public databases using transposase examples from each of the major clades and a threshold of greater than 30% identity <ref name=":0" /> indicated that members of this family are widely distributed over bacterial and archaeal genera and species. This includes the [[wikipedia:Euryarchaeota|Euryarchaeota]] [[wikipedia:Methanotorris|''Methanotorris'' ''formicicus'']] and [[wikipedia:Methanocaldococcus|''Methanocaldococcus'' ''bathoardescencs'']] and alpha-, beta-, gama- and epsilon-protobacteria, firmicutes, deinococci, cyanobacteria and actinobacteria. In total 265 new genera containing IS''982''-related elements were | + | A recent extensive [https://blast.ncbi.nlm.nih.gov/Blast.cgi BLAST] search in the non-redundant public databases using transposase examples from each of the major clades and a threshold of greater than 30% identity <ref name=":0" /> indicated that members of this family are widely distributed over bacterial and archaeal genera and species. This includes the [[wikipedia:Euryarchaeota|Euryarchaeota]] [[wikipedia:Methanotorris|''Methanotorris'' ''formicicus'']] and [[wikipedia:Methanocaldococcus|''Methanocaldococcus'' ''bathoardescencs'']] and alpha-, beta-, gama- and epsilon-protobacteria, firmicutes, deinococci, cyanobacteria and actinobacteria. In total 265 new genera containing IS''982''-related elements were ideidentified,though the IS copies from this analysis were not further characterized <ref name=":0" />. |
| + | |||
| + | Like many IS, members of this family are able to increase gene expression presumably by supplying a promoter or by formation of a hybrid promoter on insertion upstream of a resident gene. At least two members, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS982B IS''982''B] <ref><nowiki><pubmed>8602160</pubmed></nowiki></ref> and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1187 IS''1187''] [see <ref><nowiki><pubmed>PMC149317</pubmed></nowiki></ref><ref><nowiki><pubmed>23158541</pubmed></nowiki></ref><ref><nowiki><pubmed>PMC99653</pubmed></nowiki></ref>] can provide a -35 hexamer in their right '''IR''' capable of forming a hybrid promoter with a resident -10 . | ||
| − | + | A recent review of this family <ref name=":0" /> lists a number of additional elements including [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAba4 IS''Aba4''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAba9 ISA''ba9''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAba47 IS''Aba47''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAba825 IS''Aba825''] whose insertion can lead to increased [[wikipedia:Carbapenem|carbapenem]] resistance in ''[[wikipedia:Acinetobacter_baumannii|Acinetobacter baumannii]]'' <ref><nowiki><pubmed>22862829</pubmed></nowiki></ref><ref><nowiki><pubmed>PMC3028783</pubmed></nowiki></ref><ref><nowiki><pubmed>26194827</pubmed></nowiki></ref><ref><nowiki><pubmed>23143900</pubmed></nowiki></ref><ref><nowiki><pubmed>PMC6030224</pubmed></nowiki></ref><ref><nowiki><pubmed>PMC2704689</pubmed></nowiki></ref><ref><nowiki><pubmed>PMC2737879</pubmed></nowiki></ref>. In addition, as also pointed out by Fayad et al. <ref name=":0" />, insertion of members of the family which lead to gene inactivation has been observed to result in: increased antibiotic resistance ([[wikipedia:Vancomycin|vancomycin]] and [[wikipedia:Teicoplanin|teicoplanin]] by [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS19 IS''19''] <ref><nowiki><pubmed>PMC89867</pubmed></nowiki></ref> and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEfm1 IS''Efm1''] <ref><nowiki><pubmed>PMC86817</pubmed></nowiki></ref> in ''[[wikipedia:Enterococcus_faecium|Enterococcus faecium]];'' [[wikipedia:Carbapenem|carbapenem]] by [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAba825 IS''Aba825''] disruption of the CarO membrane protein <ref><nowiki><pubmed>PMC1068641</pubmed></nowiki></ref> in [[wikipedia:Acinetobacter_baumannii|''A''. ''baumannii'']]); reduced antibiotic resistance ([[wikipedia:Daptomycin|daptomycin]] in ''[[wikipedia:Enterococcus_faecium|Enterococcus faecium]];'' <ref><nowiki><pubmed>27353469</pubmed></nowiki></ref>); or in decreased virulence ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISSa4 IS''Sa4''] in ''[[wikipedia:Streptococcus_agalactiae|S. agalactiae]]'' <ref name=":1"><pubmed>10607898</pubmed> | |
| − | + | </nowiki></ref>; [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISScr1 IS''Scr1''] in ''[[wikipedia:Streptococcus|Streptococcus criceti]]'' <ref><nowiki><pubmed>22976390</pubmed></nowiki></ref>; [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS195 IS''195''] in ''[[wikipedia:Porphyromonas_gingivalis|Porphyromonas gingivalis]]'' <ref><nowiki><pubmed>PMC108310</pubmed></nowiki></ref>). | |
====Organization==== | ====Organization==== | ||
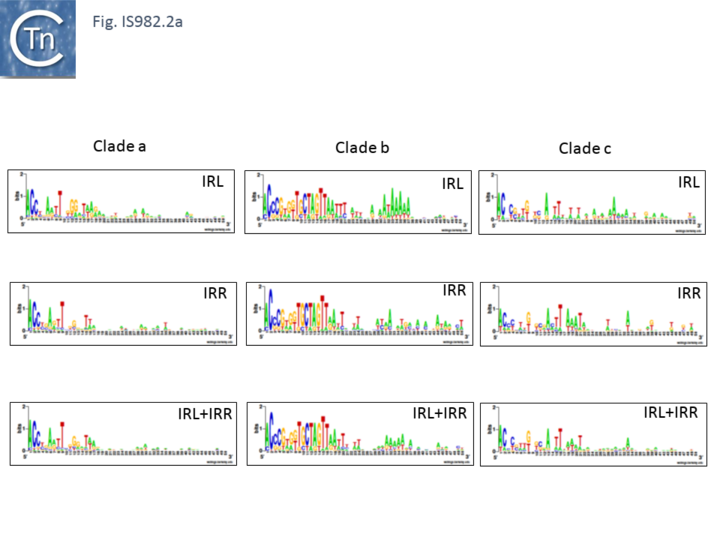
| − | IS''982'' family members are between 962 and 1155 bp long [[:File:Fig. IS982.1.png|(Fig.IS''982''.2)]], carry similar terminal IRs of between 18 and 35 bp with conserved ends: 5’-ACCC-3’ and generate DR of 6 to 8 bp e.g. ISSa''4'' from ''[[wikipedia:Streptococcus_agalactiae|Streptococcus agalactiae]]'' generates 7 bp (or perhaps 9 bp) flanking DRs <ref name=":1" /> and ISCce''1'' from [[wikipedia:Clostridium_cellulovorans|''Clostridium cellulovorans'']] is flanked by 7 bp repeats <ref><nowiki><pubmed>PMC94268</pubmed></nowiki></ref>. There is some evidence that the IR consensus varies slightly according to each IS subgroup <ref name=":0" /> [[:File:IS982.2a.png|(Fig.IS''982''.2a)]] | + | IS''982'' family members are between 962 and 1155 bp long [[:File:Fig. IS982.1.png|(Fig.IS''982''.2)]], carry similar terminal IRs of between 18 and 35 bp with conserved ends: 5’-ACCC-3’ and generate DR of 6 to 8 bp e.g. [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISSa4 ISSa''4''] from ''[[wikipedia:Streptococcus_agalactiae|Streptococcus agalactiae]]'' generates 7 bp (or perhaps 9 bp) flanking DRs <ref name=":1" /> and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISCce1 ISCce''1''] from [[wikipedia:Clostridium_cellulovorans|''Clostridium cellulovorans'']] is flanked by 7 bp repeats <ref><nowiki><pubmed>PMC94268</pubmed></nowiki></ref>. There is some evidence that the IR consensus varies slightly according to each IS subgroup <ref name=":0" /> [[:File:IS982.2a.png|(Fig.IS''982''.2a)]] |
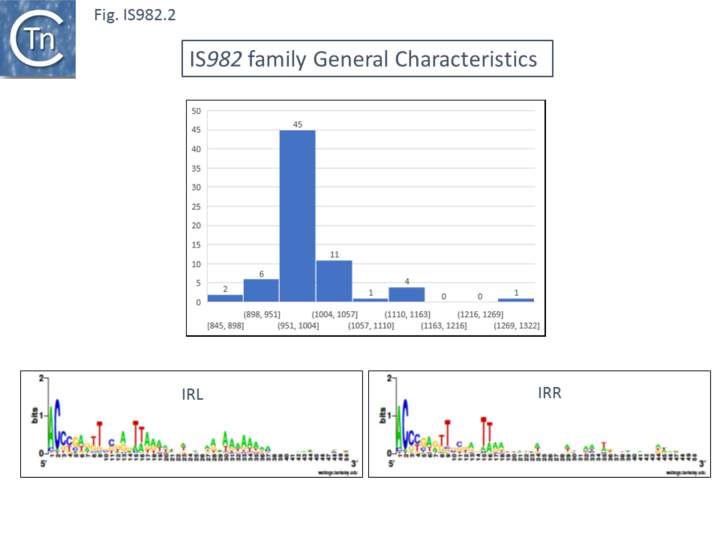
| − | [[Image:Fig. IS982.1.png|thumb|center| | + | [[Image:Fig. IS982.1.png|thumb|center|720x720px|'''Fig. IS''982''.''' '''Top:''' Distribution of IS length (base pairs) of IS''982'' family members. The number of examples used in the sample is shown above each column. '''Bottom''': Left ('''IRL''') and right '''IRR''' inverted terminal repeats are shown in WebLogo format.|alt=]] |
| + | |||
| + | [[File:IS982.2a.png|alt=|center|thumb|720x720px|'''Fig. IS''982''.2.a.''' Left ('''IRL''') inverted terminal repeats are shown in WebLogo format.]] | ||
| + | |||
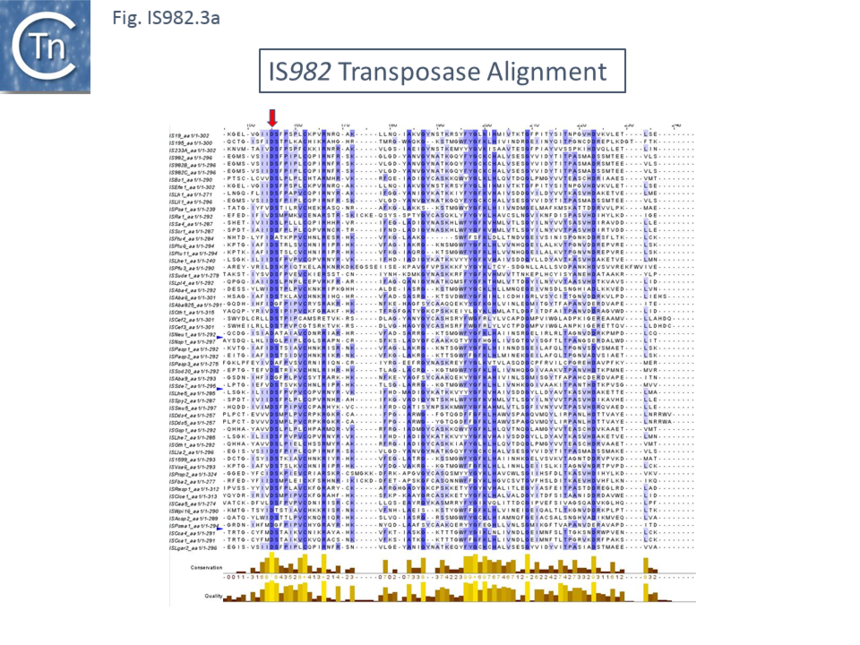
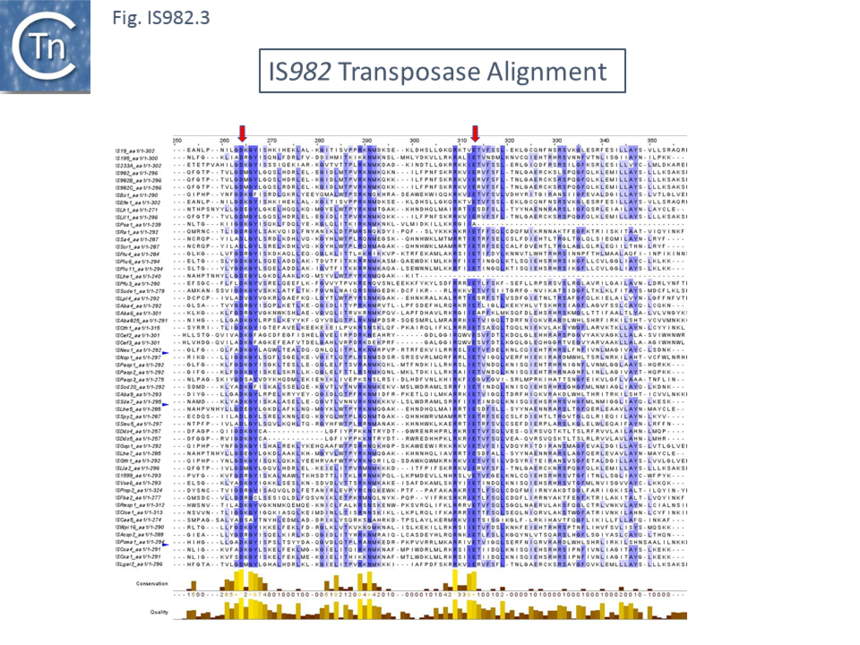
| + | Family members encode a single ''orf'' of between of 271 and 313 amino acids with a possible DDE motif but without a convincing conserved downstream K/R residue. They exhibit between 33% and 44% amino acid identity. The identities are shown in the alignment included in [[:File:Fig. IS982.1.png|(Fig.IS''982''.3 '''a''' and '''b''')]] together with the DDE motif suggested by Fayad et al. <ref name=":0" />. The N-terminal domain, which includes a potential [[wikipedia:Helix-turn-helix|HTH DNA binding motif]] <ref name=":0" /> is not shown. | ||
| + | [[File:IS982.3a.png|alt=|center|thumb|860x860px|'''Fig. IS''982''.3.a.''' Alignment of the proposed IS''982'' family transposase catalytic domain. The conserved DDE triad identified by Fayad et al <ref name=":0" /> is indicated by vertical red arrows. The N-terminal [[wikipedia:Helix-turn-helix|HTH domain]] is not shown.]] | ||
| − | |||
| − | + | Although the Tpase of a majority of members occupies a single reading phase, there are several examples in which the gene is distributed over two phases. It has been reported that a +1 nucleotide insertion in the Tpase ''orf'' of an [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISLpl4 IS''Lpl4''] from ''[[wikipedia:Oenococcus_oeni|Oenococcus oeni]]'' may undergo programmed translational frameshifting at a low rate <ref><nowiki><pubmed>16278055</pubmed></nowiki></ref>. Although this must be confirmed, it would represent the first functional case of +1 frameshifting in IS. The Tpase of archaeal element [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPfu3 IS''Pfu3''] is also distributed over two phases. [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPfu3 IS''Pfu3''] carries a potential transcriptional frameshift signal A7 (PTR) present in all five copies, suggesting that [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPfu3 IS''Pfu3''] is active. | |
| − | [ | ||
| − | + | <br /> | |
| − | [[File:IS982.3b.png|alt=|center|thumb|860x860px|'''Fig. IS''982''.3.b.''' Alignment of the proposed | + | [[File:IS982.3b.png|alt=|center|thumb|860x860px|'''Fig. IS''982''.3.b.''' Alignment of the proposed IS''982'' family transposase catalytic domain. The conserved DDE triad identified by Fayad et al <ref name=":0" /> is indicated by vertical red arrows. The N-terminal [[wikipedia:Helix-turn-helix|HTH domain]] is not shown.]] |
At present, there is no information concerning the transposition mechanism of this family. | At present, there is no information concerning the transposition mechanism of this family. | ||
==Bibliography== | ==Bibliography== | ||
<references /> | <references /> | ||
Revision as of 22:40, 11 August 2021
General
The IS982 family has over 70 entries in ISfinder from over 40 bacterial and archaeal species. In the case of ISLpl4 from Lactobacillus plantarum identical copies have been detected in Leuconostoc mesenteroides, Oenococcus oeni, and Lactobacillus sakei indicating horizontal gene transfer. Historically, a single IS982 family member, ISPfu3, was first identified in the archaea from the archaea Pyrococcus furiosus and this was later joined by ISTli1 from Thermococcus litoralis (both mrmbero of the order Thermococcales).
A phylogenetic tree of the transposase of family members (Fig. IS982.1) shows three deeply branching clades (a, b and c) which broadly correspond to those identified by Fayad et al. [1]. They can be further subdivided into several subclades (ai; aii and aii; bi, which includes streptococci and lactococci; and bii which contain bacilli and lactobacilli)
A recent extensive BLAST search in the non-redundant public databases using transposase examples from each of the major clades and a threshold of greater than 30% identity [1] indicated that members of this family are widely distributed over bacterial and archaeal genera and species. This includes the Euryarchaeota Methanotorris formicicus and Methanocaldococcus bathoardescencs and alpha-, beta-, gama- and epsilon-protobacteria, firmicutes, deinococci, cyanobacteria and actinobacteria. In total 265 new genera containing IS982-related elements were ideidentified,though the IS copies from this analysis were not further characterized [1].
Like many IS, members of this family are able to increase gene expression presumably by supplying a promoter or by formation of a hybrid promoter on insertion upstream of a resident gene. At least two members, IS982B [2] and IS1187 [see [3][4][5]] can provide a -35 hexamer in their right IR capable of forming a hybrid promoter with a resident -10 .
A recent review of this family [1] lists a number of additional elements including ISAba4, ISAba9, ISAba47 and ISAba825 whose insertion can lead to increased carbapenem resistance in Acinetobacter baumannii [6][7][8][9][10][11][12]. In addition, as also pointed out by Fayad et al. [1], insertion of members of the family which lead to gene inactivation has been observed to result in: increased antibiotic resistance (vancomycin and teicoplanin by IS19 [13] and ISEfm1 [14] in Enterococcus faecium; carbapenem by ISAba825 disruption of the CarO membrane protein [15] in A. baumannii); reduced antibiotic resistance (daptomycin in Enterococcus faecium; [16]); or in decreased virulence (ISSa4 in S. agalactiae [17]; ISScr1 in Streptococcus criceti [18]; IS195 in Porphyromonas gingivalis [19]).
Organization
IS982 family members are between 962 and 1155 bp long (Fig.IS982.2), carry similar terminal IRs of between 18 and 35 bp with conserved ends: 5’-ACCC-3’ and generate DR of 6 to 8 bp e.g. ISSa4 from Streptococcus agalactiae generates 7 bp (or perhaps 9 bp) flanking DRs [17] and ISCce1 from Clostridium cellulovorans is flanked by 7 bp repeats [20]. There is some evidence that the IR consensus varies slightly according to each IS subgroup [1] (Fig.IS982.2a)
Family members encode a single orf of between of 271 and 313 amino acids with a possible DDE motif but without a convincing conserved downstream K/R residue. They exhibit between 33% and 44% amino acid identity. The identities are shown in the alignment included in (Fig.IS982.3 a and b) together with the DDE motif suggested by Fayad et al. [1]. The N-terminal domain, which includes a potential HTH DNA binding motif [1] is not shown.
Although the Tpase of a majority of members occupies a single reading phase, there are several examples in which the gene is distributed over two phases. It has been reported that a +1 nucleotide insertion in the Tpase orf of an ISLpl4 from Oenococcus oeni may undergo programmed translational frameshifting at a low rate [21]. Although this must be confirmed, it would represent the first functional case of +1 frameshifting in IS. The Tpase of archaeal element ISPfu3 is also distributed over two phases. ISPfu3 carries a potential transcriptional frameshift signal A7 (PTR) present in all five copies, suggesting that ISPfu3 is active.
At present, there is no information concerning the transposition mechanism of this family.
Bibliography
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 </nowiki>
- ↑ <pubmed>8602160</pubmed>
- ↑ <pubmed>PMC149317</pubmed>
- ↑ <pubmed>23158541</pubmed>
- ↑ <pubmed>PMC99653</pubmed>
- ↑ <pubmed>22862829</pubmed>
- ↑ <pubmed>PMC3028783</pubmed>
- ↑ <pubmed>26194827</pubmed>
- ↑ <pubmed>23143900</pubmed>
- ↑ <pubmed>PMC6030224</pubmed>
- ↑ <pubmed>PMC2704689</pubmed>
- ↑ <pubmed>PMC2737879</pubmed>
- ↑ <pubmed>PMC89867</pubmed>
- ↑ <pubmed>PMC86817</pubmed>
- ↑ <pubmed>PMC1068641</pubmed>
- ↑ <pubmed>27353469</pubmed>
- ↑ 17.0 17.1 </nowiki>
- ↑ <pubmed>22976390</pubmed>
- ↑ <pubmed>PMC108310</pubmed>
- ↑ <pubmed>PMC94268</pubmed>
- ↑ <pubmed>16278055</pubmed>