Difference between revisions of "IS Families/IS630 family"
| Line 1: | Line 1: | ||
====Original Identification==== | ====Original Identification==== | ||
| − | IS''630'' was first identified in the ''[[wikipedia:Shigella_sonnei|Shigella sonnei]]'' genome<ref><nowiki><pubmed>2824781</pubmed></nowiki></ref>. Another transpositionally active IS''630'' family member which showed similarity to the Tc1 eukaryotic transposon family<ref name=":0"><pubmed>9305771</pubmed> | + | [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS630 IS''630''] was first identified in the ''[[wikipedia:Shigella_sonnei|Shigella sonnei]]'' genome<ref><nowiki><pubmed>2824781</pubmed></nowiki></ref>. Another transpositionally active IS''630'' family member which showed similarity to the Tc1 eukaryotic transposon family<ref name=":0"><pubmed>9305771</pubmed> |
| + | |||
| + | </nowiki></ref> has received most attention was subsequently identified in the [[wikipedia:Cyanobacteria|cyanobacterium]] ''[[wikipedia:Synechocystis|Synechocystis]]'' PCC6803 and called [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''S1987'']''<ref name=":0" />'' or [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''TcSa''] and subsequently renamed [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''Y100'']<ref name=":1"><pubmed>12193627</pubmed></nowiki></ref>. | ||
====Distribution==== | ====Distribution==== | ||
| − | There are over 200 members from over 80 bacterial and archaeal genomes. IS''630'' itself has been used to cluster subspecies of ''[[wikipedia:Aeromonas_salmonicida|Aeromonas salmonicida]]'' by high copy number IS''630'' restriction fragment length polymorphism (HCN-IS630-RFLP)<ref><nowiki><pubmed>23406017</pubmed></nowiki></ref>. | + | There are over 200 members from over 80 bacterial and archaeal genomes. [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS630 IS''630''] itself has been used to cluster subspecies of ''[[wikipedia:Aeromonas_salmonicida|Aeromonas salmonicida]]'' by high copy number [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS630 IS''630''] restriction fragment length polymorphism (HCN-[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS630 IS''630'']-RFLP)<ref><nowiki><pubmed>23406017</pubmed></nowiki></ref>. |
====Organization==== | ====Organization==== | ||
| − | Members are between 950 and 1250 bp in length with an average of 1100 bp ([[:File:Fig. IS630.2.png|Fig. IS630.2]]). They have short terminal IRs ([[:File:Fig. IS630.1.png|Fig. IS630.1]]) and generally include a single orf. However, in about nearly half of the members, the Tpase orf is distributed over two reading frames suggesting that it may be produced as a fusion protein by frameshifting. These include IS''895'' and IS''Rm2011-2'', identified several decades ago, which appear to contain two consecutive open reading frames<ref><nowiki><pubmed>1653219</pubmed></nowiki></ref>. | + | Members are between 950 and 1250 bp in length with an average of 1100 bp ([[:File:Fig. IS630.2.png|Fig. IS630.2]]). They have short terminal IRs ([[:File:Fig. IS630.1.png|Fig. IS630.1]]) and generally include a single orf. However, in about nearly half of the members, the Tpase orf is distributed over two reading frames, suggesting that it may be produced as a fusion protein by frameshifting. These include IS''895'' and IS''Rm2011-2'', identified several decades ago, which appear to contain two consecutive open reading frames<ref><nowiki><pubmed>1653219</pubmed></nowiki></ref>. |
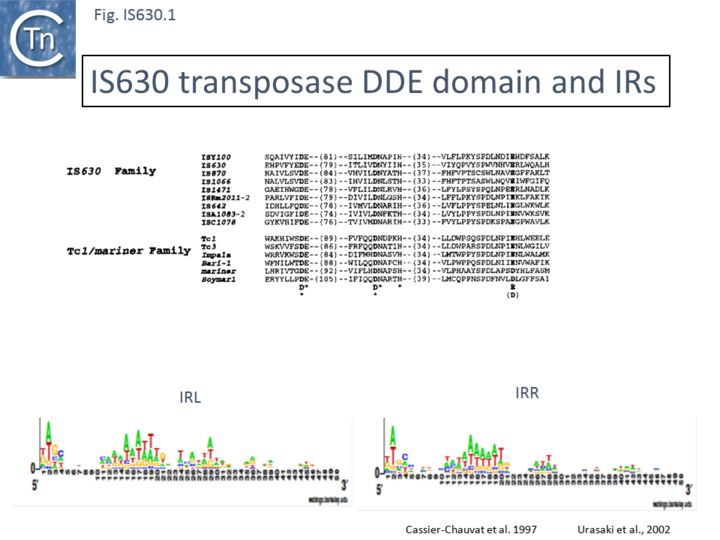
| − | [[Image:Fig. IS630.1.png|thumb|center| | + | [[Image:Fig. IS630.1.png|thumb|center|720x720px|'''Fig. IS630.1.''' '''IS''630'' DDE domains and IRs. Top''': Alignment of different IS''630'' family members and members of the Tc1/mariner family, showing the DDE domain. '''Bottom''': Left (IRL) and right IRR inverted terminal repeats are shown in WebLogo format.|alt=]] |
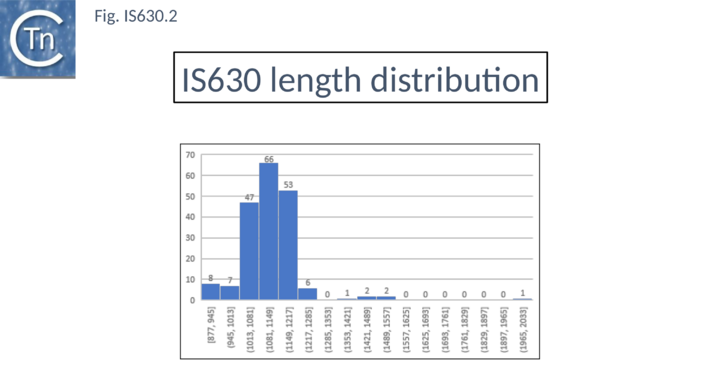
| − | [[Image:Fig. IS630.2.png|thumb|center| | + | [[Image:Fig. IS630.2.png|thumb|center|720x720px|'''Fig. IS630.2.''' '''IS630 length distribution.''' Distribution of IS length (base pairs) of IS''630'' family members. The number of examples used in the sample is shown above each column.|alt=]] |
| + | |||
| + | Other member's families carry a single long reading frame. Three elements ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS870 IS''870''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAr1 IS''Ar1''], and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISRf1 IS''Rf1'']) show more than 70% identity, two ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1066 IS''1066''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISRj1 IS''Rj1'']) show about 50% identity with each other and 40% with the other members, while two show less than 20% identity between themselves and with the other members. This is reflected in the relatively low similarity of their '''IRs''' ([[:File:Fig. IS630.1.png|Fig. IS630.1]]). | ||
| + | |||
| + | It is interesting to note that the three most closely related on the basis of Tpase similarities ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS870 IS''870''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISRf1 IS''Rf1''], and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAr1 IS''Ar1'']) appear not to carry translation termination codons for the Tpase gene. However, insertion into the specific target site, CTAG, with concomitant duplication (of either 2 or 4 base pairs) generates a TAG termination codon in phase with the Tpase gene. The influence of this arrangement on the transposition of the IS elements has yet to be determined. | ||
| + | |||
| + | Of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''Y100''] copies in the ''[[wikipedia:Synechocystis|Synechocystis]]'' PCC6803 genome, around 50% were found to include a single orf while the others included 2 smaller orfs which had been split by the presence of an additional '''A''' nucleotide in a stretch of 9 As<ref name=":1" />. While it is possible that these IS copies are inactive, it is also possible that a full-length transposase is expressed by frameshifting. Although the absence of the characteristic secondary structure signals associated with '''-1 programmed translational frameshifting''' suggests that this is unlikely, it remains possible that a transcriptional mechanism is operating. One member, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISRm2011-2 IS''Rm2011-2''] from ''[[wikipedia:Sinorhizobium_meliloti|Rhizobium meliloti]]'', also carries a [[wikipedia:Group_II_intron|group II intron]] which appears to be active ''in vivo''<ref><nowiki><pubmed>9680217</pubmed></nowiki></ref>. | ||
| − | + | The IS''630'' family is related to the Tc1/mariner family of eukaryotic TE particularly at the level of the DDE signature<ref name=":0" /> ([[:File:Fig. IS630.1.png|Fig. IS630.1]]). There is also an N-terminal [[wikipedia:Helix-turn-helix|HTH motif]]<ref name=":1" /> ([[:File:Fig. IS630.3.png|Fig. IS630.3]]) whose function in binding [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''Y100''] ends has been verified ''in vitro''<ref name=":2"><pubmed>17680987</pubmed> | |
| − | |||
| − | + | </nowiki></ref>. Moreover, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name= IS''630''] and the Tc1/mariner families target similar sequences, have similar DR and transposition of both involves cleavage of two nucleotides inside the 5’ ends<ref name=":2" /><ref name=":3"><pubmed>8556864</pubmed> | |
| − | + | </nowiki></ref>. | |
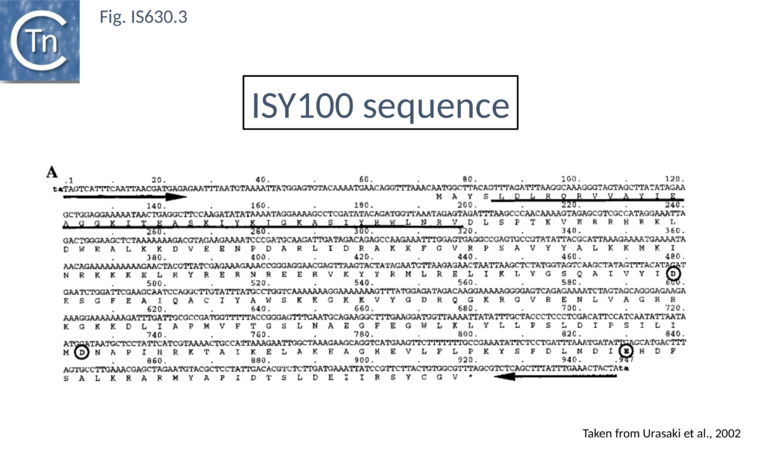
| − | [[Image:Fig. IS630.3.png|thumb|center| | + | [[Image:Fig. IS630.3.png|thumb|center|760x760px|'''Fig. IS630.3.''' The amino acid sequence of transposase encoded by [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''Y100''] is shown below the nucleotide sequence. The TA sequences adjacent to both ends of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''Y100''] sequence indicate the target site sequence duplicated on the transposition of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''Y100'']. The arrows show the terminal '''IRs''', '''IRL''' and '''IRR'''. The underlined amino acid sequence is the possible DNA-binding region with a helix-turn-helix motif. Amino acid residues of the DDE motif are circled.|alt=]] |
| − | Analysis of the ''[[wikipedia:Streptococcus_pneumoniae|Streptococcus pneumoniae]]'' genome revealed several hundred copies of short imperfect palindromic sequences (RUPs) whose ends show strong homology to those of a full length putative IS''630''-related element also present in the genome<ref><nowiki><pubmed>10537186</pubmed></nowiki></ref>. Comparison of empty and full sites from different ''[[wikipedia:Streptococcus_pneumoniae|S. pneumoniae]]'' strains indicated that, like most full-length members of the family, the RUPs are flanked by a TA dinucleotide target repeat. Although transposition of RUP sequences has yet to be demonstrated, they are structurally similar to the IS''231''-related MIC231 elements in ''[[wikipedia:Bacillus_thuringiensis|B. thuringiensis]]'' (see "[[IS Families/IS4 and related families|IS''4'' family]]") and to several eukaryote systems where many truncated copies of an element may exist together with a full length functional copy that is capable of complementing the non-autonomous copies to drive their transposition. Other IS''630''-based MITES have been identified in ''[[wikipedia:Escherichia_coli|Escherichia coli]]'', ''[[wikipedia:Photorhabdus_luminescens|Photorhabdus luminescens]]''<ref><nowiki><pubmed>14528314</pubmed></nowiki></ref> and ''[[wikipedia:Yersinia_pestis|Yersinia pestis]]''<ref><nowiki><pubmed>16963573</pubmed></nowiki></ref>. | + | Analysis of the ''[[wikipedia:Streptococcus_pneumoniae|Streptococcus pneumoniae]]'' genome revealed several hundred copies of short imperfect palindromic sequences (RUPs) whose ends show strong homology to those of a full length putative [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS630 IS''630'']-related element also present in the genome<ref><nowiki><pubmed>10537186</pubmed></nowiki></ref>. Comparison of empty and full sites from different ''[[wikipedia:Streptococcus_pneumoniae|S. pneumoniae]]'' strains indicated that, like most full-length members of the family, the RUPs are flanked by a TA dinucleotide target repeat. Although transposition of RUP sequences has yet to be demonstrated, they are structurally similar to the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS231 IS''231'']-related MIC231 elements in ''[[wikipedia:Bacillus_thuringiensis|B. thuringiensis]]'' (see "[[IS Families/IS4 and related families|IS''4'' family]]") and to several eukaryote systems where many truncated copies of an element may exist together with a full length functional copy that is capable of complementing the non-autonomous copies to drive their transposition. Other [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS630 IS''630'']-based MITES have been identified in ''[[wikipedia:Escherichia_coli|Escherichia coli]]'', ''[[wikipedia:Photorhabdus_luminescens|Photorhabdus luminescens]]''<ref><nowiki><pubmed>14528314</pubmed></nowiki></ref> and ''[[wikipedia:Yersinia_pestis|Yersinia pestis]]''<ref><nowiki><pubmed>16963573</pubmed></nowiki></ref>. |
====Insertion Specificity==== | ====Insertion Specificity==== | ||
| − | Family members show high target specificity inserting into and duplicating a TA dinucleotide with a preference for the sequence 5’-NTAN-3’<ref name=":4"><pubmed>1655702</pubmed> | + | Family members show high target specificity inserting into and duplicating a TA dinucleotide, with a preference for the sequence 5’-NTAN-3’<ref name=":4"><pubmed>1655702</pubmed> |
| + | |||
| + | </nowiki></ref>. Since the cleavages of the non-transferred strand occur 2 nts within the 5’ end of the IS, repair of the donor molecule after excision of the IS can result in a 2 bp scar at the excision site. | ||
| + | |||
| + | Detailed studies concerning the target DNA sequence have been carried out in the case of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS630 IS''630'']<ref name=":5"><pubmed>2163390</pubmed> | ||
| − | + | </nowiki></ref>. The target sequence was determined before and after insertion, and the results suggested that insertion generated a duplication of an invariant target TA dinucleotide. That this dinucleotide does not form an integral part of the IS was investigated by site-specific mutagenesis of the transposon donor to eliminate the terminal TA. Transposition of the IS from the mutated donor molecule resulted in insertions which all exhibited a flanking TA direct repeat. This clearly demonstrates that insertion results in the duplication of the central TA dinucleotide<ref name=":5" />. Further analysis demonstrated that [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS630 IS''630''] exhibits a strong preference for a 5'-CTAG-3' target sequence<ref name=":4" />. Point mutation of the CTAG target sites reduced or eliminated their attractiveness as insertion hotspots. The two preferred insertion sites were identified in plasmid ColE1 corresponded to TA sequences in the inverted repeats of a 13-base-pair stem region of the [[wikipedia:Rho_factor|[rho]-dependent transcription terminator]]. IS''630'' is flanked by TA, and ''in vitro'' mutagenesis of the flanking TA did not affect further transposition activity, the ability to insert preferentially into the TA within the 13-base-pair inverted repeat or to duplicate its target sequence<ref name=":4" /><ref name=":5" />. | |
| − | All known insertions of these elements are consistent with a TA duplication. In two insertions of the ''[[wikipedia:Allorhizobium_vitis|Agrobacterium vitis]]'' element IS''870'' it was possible to determine the target sequence prior to insertion. This was found to carry a single CTAG copy, an observation that remains consistent with a simple TA dinucleotide target duplication. We assume that all members of this family generate an identical (TA) target duplication and the tips of the elements have therefore been defined accordingly in [https://isfinder.biotoul.fr/ ISfinder]. | + | All known insertions of these elements are consistent with a TA duplication. In two insertions of the ''[[wikipedia:Allorhizobium_vitis|Agrobacterium vitis]]'' element [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS870 IS''870''] it was possible to determine the target sequence prior to insertion. This was found to carry a single CTAG copy, an observation that remains consistent with a simple TA dinucleotide target duplication. We assume that all members of this family generate an identical (TA) target duplication, and the tips of the elements have therefore been defined accordingly in [https://isfinder.biotoul.fr/ ISfinder]. |
====Mechanism==== | ====Mechanism==== | ||
| − | ''In vivo'' studies <ref name=":1" /> demonstrated that, when supplied with transposase, the IS''630'' family member, IS''Y100'' (IS''TcSa'') first identified in ''[[wikipedia:Synechocystis|Synechocystis]]'' sp. PCC6803<ref name=":0" /> generates linear forms which terminate exactly at the 3’ ends but which lack 2 nucleotides at the 5’ ends resulting in a 2 base overhang. Both ''in vivo <ref name=":1" />'' and ''in vitro'' using circular plasmid DNA<ref name=":2" />. This is also a characteristic of transposition of the eukaryote Tc1 transposon<ref><nowiki><pubmed>10431195</pubmed></nowiki></ref>. Although IS circles had been observed, as for Tc1 these are thought to be dead-end products and not transposition intermediates<ref name=":1" />. | + | ''In vivo'' studies <ref name=":1" /> demonstrated that, when supplied with transposase, the IS''630'' family member, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''Y100''] ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''TcSa'']) first identified in ''[[wikipedia:Synechocystis|Synechocystis]]'' sp. PCC6803<ref name=":0" /> generates linear forms which terminate exactly at the 3’ ends but which lack 2 nucleotides at the 5’ ends resulting in a 2 base overhang. Both ''in vivo <ref name=":1" />'' and ''in vitro'' using circular plasmid DNA<ref name=":2" />. This is also a characteristic of transposition of the eukaryote Tc1 transposon<ref><nowiki><pubmed>10431195</pubmed></nowiki></ref>. Although IS circles had been observed, as for Tc1 these are thought to be dead-end products and not transposition intermediates<ref name=":1" />. |
| − | In the case of IS''630'' itself, while direct insertions can be observed at reasonable frequencies, no cointegrates could be detected<ref name=":5" />. | + | In the case of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS630 IS''630''] itself, while direct insertions can be observed at reasonable frequencies, no cointegrates could be detected<ref name=":5" />. |
| − | IS''630'' transposition has been addressed ''in vitro'' using IS''Y100<ref name=":2" />''. The Tpase was shown to specifically bind IS''Y100'' IR using an N-terminal domain containing two potential [[wikipedia:Helix-turn-helix|HTH motifs]]. It is the only protein required for IS''Y100'' excision and integration and introduces double-strand breaks on mini-IS''Y100'' on a supercoiled DNA substrate. Tc1/mariner element transposition has also been extensively studied ''in vitro<ref name=":3" />'' and a Tpase structural model is available [e.g. <ref><nowiki><pubmed>12535535</pubmed></nowiki></ref><ref><nowiki><pubmed>16511570</pubmed></nowiki></ref>]. IS''630'' Tpase cleaves exactly at the 3’ (transferred strand) IS ends and two nucleotides inside the 5’ (non-transferred strand) ends. Cleavage is less precise on linear substrates. Both single-end and, less frequently, double-end insertion occur ''in vitro'' in a TA-target-specific manner<ref name=":6"><pubmed>19965773</pubmed> | + | IS''630'' transposition has been addressed ''in vitro'' using [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''Y100'']''<ref name=":2" />''. The Tpase was shown to specifically bind IS''Y100'' IR using an N-terminal domain containing two potential [[wikipedia:Helix-turn-helix|HTH motifs]]. It is the only protein required for [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''Y100''] excision and integration and introduces double-strand breaks on mini-[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''Y100''] on a supercoiled DNA substrate. Tc1/mariner element transposition has also been extensively studied ''in vitro<ref name=":3" />'' and a Tpase structural model is available [e.g. <ref><nowiki><pubmed>12535535</pubmed></nowiki></ref><ref><nowiki><pubmed>16511570</pubmed></nowiki></ref>]. [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS630 IS''630''] Tpase cleaves exactly at the 3’ (transferred strand) IS ends and two nucleotides inside the 5’ (non-transferred strand) ends. Cleavage is less precise on linear substrates. Both single-end and, less frequently, double-end insertion occur ''in vitro'' in a TA-target-specific manner<ref name=":6"><pubmed>19965773</pubmed></nowiki></ref>. Transposition does not involve a hairpin intermediate. |
| − | The related eukaryote Tc1 element is known to transpose by a cut and paste mechanism and leaves a “footprint” (additional bases in the original donor site) on excision ([[:File:Fig. IS630.4.png|Fig. IS630.4]]). When purified vector backbone produced by transposase mediated cleavage of IS''Y100'' ''in vitro'' was circularised ''In vitro'', gel- produced by transposase-mediated cleavage of pISY100 -kan was efficiently circularized with T4 DNA ligase the predicted TATATA (i.e. a footprint) junction sequence was recovered after transformation into ''E. coli''. | + | The related eukaryote Tc1 element is known to transpose by a '''cut-and-paste mechanism''' and leaves a “footprint” (additional bases in the original donor site) on excision ([[:File:Fig. IS630.4.png|Fig. IS630.4]]). When purified vector backbone produced by transposase mediated cleavage of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''Y100''] ''in vitro'' was circularised ''In vitro'', gel- produced by transposase-mediated cleavage of pISY100 -kan was efficiently circularized with T4 DNA ligase the predicted TATATA (i.e. a footprint) junction sequence was recovered after transformation into ''E. coli''. |
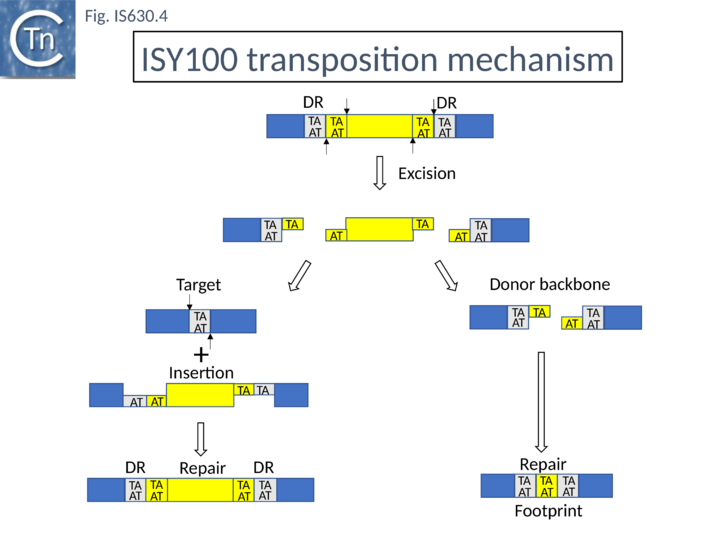
| − | [[Image:Fig. IS630.4.png|thumb|center| | + | [[Image:Fig. IS630.4.png|thumb|center|720x720px|'''Fig. IS630.4. Proposed IS''630'' ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''Y100'']) Transposition Pathway.''' From Urasaki et al (2002) and from Plasterk et al., (1999) for the related element Tc1/mariner Transposase binds to the IS (yellow) ends catalyzes staggered double-strand breaks at each end (one to generate a 3’OH at the IS tip and the second, 2 bases within the IS to generate a recessed 5’ end), excising the IS from its donor site (blue). The 3’OH group then attacks the conserved TA dinucleotide target. Host enzymes then repair the ensuing gaps (left). In this model, excision and repair result in the introduction of a small repetition at the donor site, called a “footprint” in the case of Tc1/mariner. The model has been confirmed by in vitro studies (Feng and Colloms, 2007).|alt=]] |
| − | It has also been demonstrated that fusion of a zinc-finger DNA-binding domain of Zif268 to the IS''Y100'' transposase C-terminus targets integration into TA dinucleotides positioned 6-17 bp to one side of a Zif268 binding site. The targeting specificity can be changed with Zif268 variants which recognize other sequences<ref name=":6" />. | + | It has also been demonstrated that fusion of a zinc-finger DNA-binding domain of Zif268 to the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISTcSa IS''Y100''] transposase C-terminus targets integration into TA dinucleotides positioned 6-17 bp to one side of a Zif268 binding site. The targeting specificity can be changed with Zif268 variants which recognize other sequences<ref name=":6" />. |
<br /> | <br /> | ||
==Bibliography== | ==Bibliography== | ||
<references /> | <references /> | ||
Revision as of 22:19, 11 August 2021
Contents
Original Identification
IS630 was first identified in the Shigella sonnei genome[1]. Another transpositionally active IS630 family member which showed similarity to the Tc1 eukaryotic transposon family[2] has received most attention was subsequently identified in the cyanobacterium Synechocystis PCC6803 and called ISS1987[2] or ISTcSa and subsequently renamed ISY100[3].
Distribution
There are over 200 members from over 80 bacterial and archaeal genomes. IS630 itself has been used to cluster subspecies of Aeromonas salmonicida by high copy number IS630 restriction fragment length polymorphism (HCN-IS630-RFLP)[4].
Organization
Members are between 950 and 1250 bp in length with an average of 1100 bp (Fig. IS630.2). They have short terminal IRs (Fig. IS630.1) and generally include a single orf. However, in about nearly half of the members, the Tpase orf is distributed over two reading frames, suggesting that it may be produced as a fusion protein by frameshifting. These include IS895 and ISRm2011-2, identified several decades ago, which appear to contain two consecutive open reading frames[5].
Other member's families carry a single long reading frame. Three elements (IS870, ISAr1, and ISRf1) show more than 70% identity, two (IS1066 and ISRj1) show about 50% identity with each other and 40% with the other members, while two show less than 20% identity between themselves and with the other members. This is reflected in the relatively low similarity of their IRs (Fig. IS630.1).
It is interesting to note that the three most closely related on the basis of Tpase similarities (IS870, ISRf1, and ISAr1) appear not to carry translation termination codons for the Tpase gene. However, insertion into the specific target site, CTAG, with concomitant duplication (of either 2 or 4 base pairs) generates a TAG termination codon in phase with the Tpase gene. The influence of this arrangement on the transposition of the IS elements has yet to be determined.
Of the ISY100 copies in the Synechocystis PCC6803 genome, around 50% were found to include a single orf while the others included 2 smaller orfs which had been split by the presence of an additional A nucleotide in a stretch of 9 As[3]. While it is possible that these IS copies are inactive, it is also possible that a full-length transposase is expressed by frameshifting. Although the absence of the characteristic secondary structure signals associated with -1 programmed translational frameshifting suggests that this is unlikely, it remains possible that a transcriptional mechanism is operating. One member, ISRm2011-2 from Rhizobium meliloti, also carries a group II intron which appears to be active in vivo[6].
The IS630 family is related to the Tc1/mariner family of eukaryotic TE particularly at the level of the DDE signature[2] (Fig. IS630.1). There is also an N-terminal HTH motif[3] (Fig. IS630.3) whose function in binding ISY100 ends has been verified in vitro[7]. Moreover, IS630 and the Tc1/mariner families target similar sequences, have similar DR and transposition of both involves cleavage of two nucleotides inside the 5’ ends[7][8].
Analysis of the Streptococcus pneumoniae genome revealed several hundred copies of short imperfect palindromic sequences (RUPs) whose ends show strong homology to those of a full length putative IS630-related element also present in the genome[9]. Comparison of empty and full sites from different S. pneumoniae strains indicated that, like most full-length members of the family, the RUPs are flanked by a TA dinucleotide target repeat. Although transposition of RUP sequences has yet to be demonstrated, they are structurally similar to the IS231-related MIC231 elements in B. thuringiensis (see "IS4 family") and to several eukaryote systems where many truncated copies of an element may exist together with a full length functional copy that is capable of complementing the non-autonomous copies to drive their transposition. Other IS630-based MITES have been identified in Escherichia coli, Photorhabdus luminescens[10] and Yersinia pestis[11].
Insertion Specificity
Family members show high target specificity inserting into and duplicating a TA dinucleotide, with a preference for the sequence 5’-NTAN-3’[12]. Since the cleavages of the non-transferred strand occur 2 nts within the 5’ end of the IS, repair of the donor molecule after excision of the IS can result in a 2 bp scar at the excision site.
Detailed studies concerning the target DNA sequence have been carried out in the case of IS630[13]. The target sequence was determined before and after insertion, and the results suggested that insertion generated a duplication of an invariant target TA dinucleotide. That this dinucleotide does not form an integral part of the IS was investigated by site-specific mutagenesis of the transposon donor to eliminate the terminal TA. Transposition of the IS from the mutated donor molecule resulted in insertions which all exhibited a flanking TA direct repeat. This clearly demonstrates that insertion results in the duplication of the central TA dinucleotide[13]. Further analysis demonstrated that IS630 exhibits a strong preference for a 5'-CTAG-3' target sequence[12]. Point mutation of the CTAG target sites reduced or eliminated their attractiveness as insertion hotspots. The two preferred insertion sites were identified in plasmid ColE1 corresponded to TA sequences in the inverted repeats of a 13-base-pair stem region of the [rho]-dependent transcription terminator. IS630 is flanked by TA, and in vitro mutagenesis of the flanking TA did not affect further transposition activity, the ability to insert preferentially into the TA within the 13-base-pair inverted repeat or to duplicate its target sequence[12][13].
All known insertions of these elements are consistent with a TA duplication. In two insertions of the Agrobacterium vitis element IS870 it was possible to determine the target sequence prior to insertion. This was found to carry a single CTAG copy, an observation that remains consistent with a simple TA dinucleotide target duplication. We assume that all members of this family generate an identical (TA) target duplication, and the tips of the elements have therefore been defined accordingly in ISfinder.
Mechanism
In vivo studies [3] demonstrated that, when supplied with transposase, the IS630 family member, ISY100 (ISTcSa) first identified in Synechocystis sp. PCC6803[2] generates linear forms which terminate exactly at the 3’ ends but which lack 2 nucleotides at the 5’ ends resulting in a 2 base overhang. Both in vivo [3] and in vitro using circular plasmid DNA[7]. This is also a characteristic of transposition of the eukaryote Tc1 transposon[14]. Although IS circles had been observed, as for Tc1 these are thought to be dead-end products and not transposition intermediates[3].
In the case of IS630 itself, while direct insertions can be observed at reasonable frequencies, no cointegrates could be detected[13].
IS630 transposition has been addressed in vitro using ISY100[7]. The Tpase was shown to specifically bind ISY100 IR using an N-terminal domain containing two potential HTH motifs. It is the only protein required for ISY100 excision and integration and introduces double-strand breaks on mini-ISY100 on a supercoiled DNA substrate. Tc1/mariner element transposition has also been extensively studied in vitro[8] and a Tpase structural model is available [e.g. [15][16]]. IS630 Tpase cleaves exactly at the 3’ (transferred strand) IS ends and two nucleotides inside the 5’ (non-transferred strand) ends. Cleavage is less precise on linear substrates. Both single-end and, less frequently, double-end insertion occur in vitro in a TA-target-specific manner[17]. Transposition does not involve a hairpin intermediate.
The related eukaryote Tc1 element is known to transpose by a cut-and-paste mechanism and leaves a “footprint” (additional bases in the original donor site) on excision (Fig. IS630.4). When purified vector backbone produced by transposase mediated cleavage of ISY100 in vitro was circularised In vitro, gel- produced by transposase-mediated cleavage of pISY100 -kan was efficiently circularized with T4 DNA ligase the predicted TATATA (i.e. a footprint) junction sequence was recovered after transformation into E. coli.
It has also been demonstrated that fusion of a zinc-finger DNA-binding domain of Zif268 to the ISY100 transposase C-terminus targets integration into TA dinucleotides positioned 6-17 bp to one side of a Zif268 binding site. The targeting specificity can be changed with Zif268 variants which recognize other sequences[17].
Bibliography
- ↑ <pubmed>2824781</pubmed>
- ↑ 2.0 2.1 2.2 2.3 Cassier-Chauvat C, Poncelet M, Chauvat F . Three insertion sequences from the cyanobacterium Synechocystis PCC6803 support the occurrence of horizontal DNA transfer among bacteria. - Gene: 1997 Aug 22, 195(2);257-66 [PubMed:9305771] [DOI] </nowiki>
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Urasaki A, Sekine Y, Ohtsubo E . Transposition of cyanobacterium insertion element ISY100 in Escherichia coli. - J Bacteriol: 2002 Sep, 184(18);5104-12 [PubMed:12193627] [DOI] </nowiki>
- ↑ <pubmed>23406017</pubmed>
- ↑ <pubmed>1653219</pubmed>
- ↑ <pubmed>9680217</pubmed>
- ↑ 7.0 7.1 7.2 7.3 Feng X, Colloms SD . In vitro transposition of ISY100, a bacterial insertion sequence belonging to the Tc1/mariner family. - Mol Microbiol: 2007 Sep, 65(6);1432-43 [PubMed:17680987] [DOI] </nowiki>
- ↑ 8.0 8.1 Plasterk RH . The Tc1/mariner transposon family. - Curr Top Microbiol Immunol: 1996, 204;125-43 [PubMed:8556864] [DOI] </nowiki>
- ↑ <pubmed>10537186</pubmed>
- ↑ <pubmed>14528314</pubmed>
- ↑ <pubmed>16963573</pubmed>
- ↑ 12.0 12.1 12.2 Tenzen T, Ohtsubo E . Preferential transposition of an IS630-associated composite transposon to TA in the 5'-CTAG-3' sequence. - J Bacteriol: 1991 Oct, 173(19);6207-12 [PubMed:1655702] [DOI] </nowiki>
- ↑ 13.0 13.1 13.2 13.3 Tenzen T, Matsutani S, Ohtsubo E . Site-specific transposition of insertion sequence IS630. - J Bacteriol: 1990 Jul, 172(7);3830-6 [PubMed:2163390] [DOI] </nowiki>
- ↑ <pubmed>10431195</pubmed>
- ↑ <pubmed>12535535</pubmed>
- ↑ <pubmed>16511570</pubmed>
- ↑ 17.0 17.1 Feng X, Bednarz AL, Colloms SD . Precise targeted integration by a chimaeric transposase zinc-finger fusion protein. - Nucleic Acids Res: 2010 Mar, 38(4);1204-16 [PubMed:19965773] [DOI] </nowiki>