Transposons families/Tn7 family
Contents
- 1 Historical
- 2 The Unique Chromosome insertion site attTn7
- 3 Tn7 Organization
- 4 Tn7-encoded Proteins
- 5 Tn7 Function
- 6 AttTn7, the Role of TnsD and Damage Control
- 7 In vitro transposition
- 8 A Model for TnsC/D attTn7 Interactions.
- 9 Triplex DNA as a Target
- 10 Host Factors in Tn7 Transposition
- 11 Strand Transfer Reactions of Tn7 Ends.
- 12 Importance of the Teminal Dinucleotide
- 13 Donor Strand Cleavage Reactions of Tn7 Ends.
- 14 The TnsA-TnsB transposase complex and a switch between cut-and-paste and Replicative Transposition.
- 15 TnsA and TnsB Form a Heteromeric Transposase
- 16 Effect of the divalent metal ion on cleavage activity
- 17 A Minimal Tn7 Recombination System Using Only TnsA and TnsB
- 18 Functional analysis of Tn7 Ends
- 19 Repression of Expression from the Transposase Promoter
- 20 Identification of TnsB Binding Sites
- 21 TnsB Structure in Complex with Tn7R DNA
- 22 TnsA Resembles a type IIS Restriction Endonuclease
- 23 Protease foot printing of TnsA
- 24 The TnsC C-Terminus Interacts with TnsA and TnsB
- 25 TnsC Structure.
- 26 The role of TnsE
- 26.1 Insertion Proximal to the E. coli Replication Terminus
- 26.2 Targeting to Double Strand DNA Breaks
- 26.3 Insertion into Conjugating Plasmids
- 26.4 A mechanism to promote transposon dispersion.
- 26.5 TnsABC+E Pathway Targets Replication Forks.
- 26.6 TnsE Interacts Directly with 3’ Recessed Duplex DNA
- 26.7 TnsE also Interacts with the Replication Processivity Factor DnaN
- 26.8 TnsABC+E in vitro Transposition and the Effect of the β clamp
- 27 Immunity
- 28 Tn7-related CAST Transposons
- 29 Tn7 Integron
- 30 Tn7 Family Diversity
- 31 Bibliography
Historical
Plasmid R483 was first suspected to carry a trimethoprim and streptomycin resistance transposon because these plasmid determinants became genetically linked to and transferred by a second plasmid, R144, in the host strain [1]. That the two resistance genes were part of a transposable element was confirmed by showing that they could be transposed from the incI→ plasmid, R483, to plasmids of other incompatibility groups such as R391(IncJ), R7K(IncW), and RP4(IncP), which all acquired a DNA segment of about 9 x 106 daltons as judged by sucrose density centrifugation [2]. Remarkably, in contrast to other transposons at the time, transposition into the host E. coli chromosome was also observed to a single locus between dnaA and ilv [2]. The authors pointed out that insertion did not suppress a dnaAts phenotype indicating that it did not involve the entire R483 plasmid. This is because an integrated plasmid was known to suppress dnaAts thermosensitivity by providing an alternative (integrated plasmid-driven) origin of replication for the chromosome [3][4][5], a phenomenon known as integrative suppression [6]. The chromosomally located resistance determinant could undergo subsequent transposition to another plasmid target in a recA independent manner. The transposable resistance determinant was given the name TnC [2] which later became Tn7 [7][8].
The Unique Chromosome insertion site attTn7
The nature of the specific chromosomal insertion site was investigated further [9]: by conjugation, where it was shown to be tightly genetically linked to the chromosomal markers glmS and ilv; by restriction enzyme mapping and hybridization, where it was shown to have integrated into the chromosome at the same unique position in a number of independent “transposants”; and by cloning the chromosome region into a suitable vector plasmid where it was shown that the chromosomal fragment acted as an efficient Tn7 transposition target when cloned in either orientation and, moreover, that Tn7 insertion always occurred in the same orientation with respect to the cloned fragment.
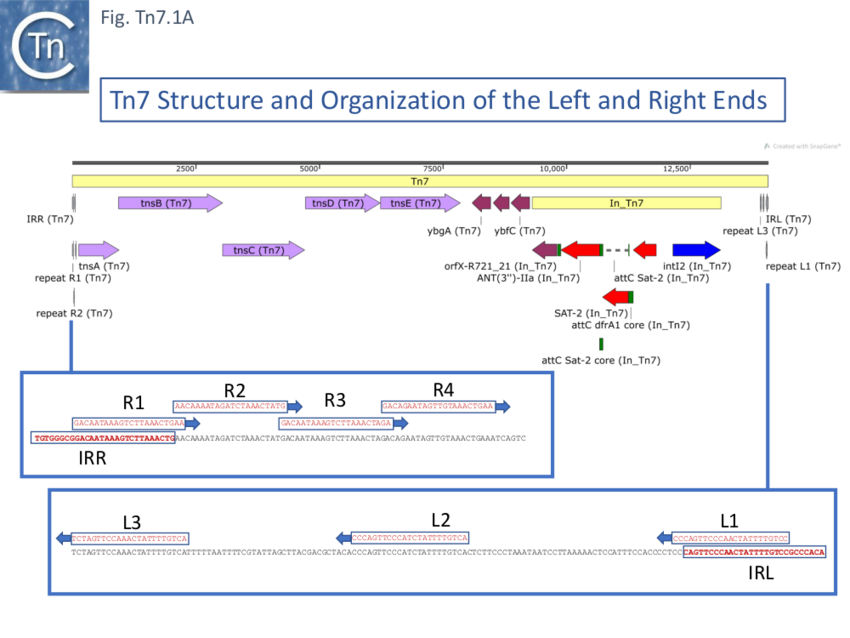
Further analysis of the chromosomal insertion site (which the authors called attTn7 in analogy to bacteriophage attB sites) by DNA sequence analysis [10] identified the junction between the target and each Tn7 end and revealed that insertion created a 5bp direct target repeat and that the ends exhibited short terminal inverted repeats but contained, 22bp repeated sequences over a longer distance which were asymmetric at each end (Fig. Tn7.1A) (see also [11]). These were contiguous in the right end but were separated at the left end [10]. It was suggested that this asymmetry may play a role in the unique insertion orientation of Tn7 in the attachment site. Note that for historical reasons, in contrast to other transposons, the left and right transposon ends are inversed for Tn7. (That is: the end proximal to the transposition genes is labelled Tn7R whereas in most other transposition systems the convention is that the transposase proximal end is defined as IRL). Later studies revealed that 199 bp of Tn7R and 166 bp of Tn7L were sufficient for transposition activity when transposition functions were provided in trans [12].
Insertion Occurs Downtream from glmS in the Terminator
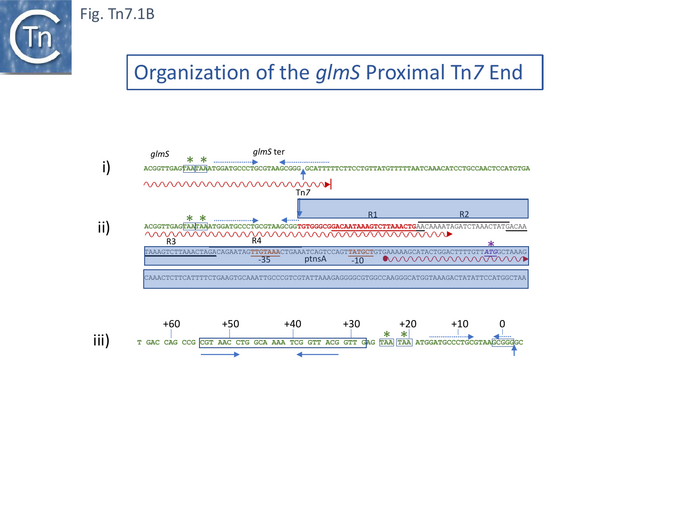
A detailed investigation into the nature of the chromosomal insertion site [13] showed it to be the glmS transcriptional terminator. Tn7 insertion abolishes glmS transcription termination permitting transcription to continue 127 base pairs into Tn7R (Fig. Tn7.1B). This study found several minor differences in the previously described sequence [9] probably due to the technical problems of sequencing at the time. The authors suggested that the insertion provides a coupling between Tn7 transposition and host growth. The IRR end carries a reading frame with showed similarities to the transposases of IS1 and Tn3 and a weak promoter and it was proposed that transcripts from glmS may regulate Tn7 transposase expression by promoter occlusion [13]. It should be noted that while Tn7 insertion inactivates the glmS transcription termination signal, it leaves the glmS gene itself intact. Moreover, it was shown that transcription across attTn7 from the glmS direction, reduces the frequency of insertion [14].
The Insertion site is Conserved in Many Bacterial Species.
Tn7 was initially used quite extensively for transposition mutagenesis and mapping before its organisation and transposition functions had been addressed. These applications included construction of genetic maps of RP4 [15][16] and R91-5 [17] and probing the organisation of the Agrobacterium tumefaciens Ti plasmid [18]. Its use was restricted to plasmids due to its sequence and orientation specific insertion not only into the E. coli chromosome but into chromosomes of other bacteria such as Vibrio [19][20], Pseudomonas [21], Caulobacter [22] or Thiobacillus [23]. However, one report concluded that although Tn7 inserts into a single chromosome locus of Pasteurella multocida, not only does it insert in both orientation but also apparently generates cointegrates [24], an observation which warrants further investigation.
However, although Tn7 inserts principally into attTn7, it was observed to be capable of insertion into other chromosomal sites but at a lower frequency [15][16].
It was also noted throughout these early studies, that Tn7 does not appear to generate cointegrates [25][26] but see [24] and “A switch between cut-and-paste and Replicative Transposition” below.
Tn7 Organization
A Tn7 restriction map [27] placed the drug resistance genes to one side of a unique EcoRI cleavage site and revealed that deletion of some DNA on the other side resulted in a derivative unable to transpose unless complemented by a wildtype Tn7 (see Fig. Tn7.1Ci). Finer deletion mapping generated from a Tn7 derivative with an IS1 insertion [25] defined two segments essential in cis for transposition (the Tn7 ends) (see also [28]), the location of the trimethoprim resistance gene (Tp), the streptomycin/spectinomycin (Sm/Sp) resistance determinant and three regions which contain Tn7 transposition functions (tnp7A, tnp7B and tnp7C) and which cover the Tn7 transposition genes known today (Fig. Tn7.1Cii).
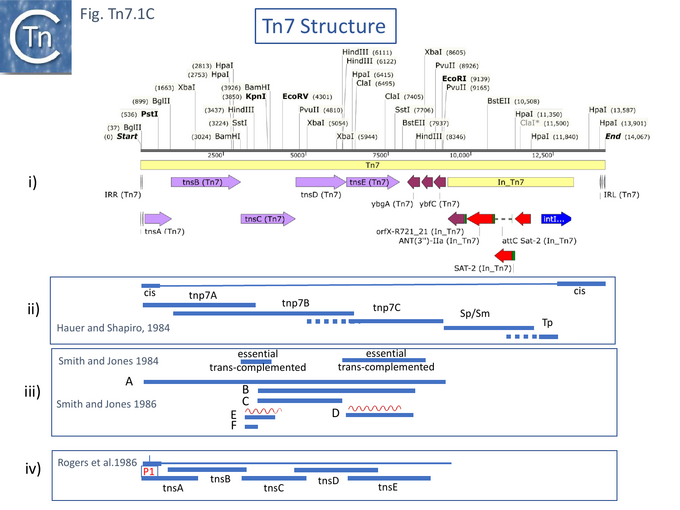
Another deletion study [26] observed that two segments, 2.1kb HindIII (fragment D) and 0.9kb BamHI (fragment E) (Fig. Tn7.1Ciii), were essential for transposition but deletions could be complemented in trans, and, moreover, when cloned alone, the HindIII fragment strongly stimulated transposition of another plasmid-carried Tn7. This segment encodes what is now known as TnsE. Additionally, Smith and Jones [29] obtained the DNA sequence of this 2.1kb HindIII fragment and identified an open reading frame encoding a protein of 538 amino acids with a GTG start codon and visualised its protein product. They also identified a second open reading frame which initiated in fragment E and continued into the next segment. This is now known to be TnsC. From the behavior of the two gene products from these two fragments, they suggested that they must interact with each other. Moreover, using appropriate transcription/translation fusions involving the chloramphenicol acetyltransferase and β-galactosidase genes, they identified a promoter located to the left of fragment E and a transcription terminator within fragment F.
Tn7-encoded Proteins
In another study [30] six Tn7-encoded proteins were identified (p18, p32, p40, p54, p85-a and p85-b,labelled according to their apparent molecular weight). These included two passenger genes (p18, a type I dihydrofolate reductase – trimethoprim resistance - and p32, the probable streptomycin/spectinomycin resistance protein, although additional passenger genes have since been identified) and four proteins involved in transposition (p40, p54, p85-a and p85-b). Both passenger genes mapped to the right-hand part of Tn7 (as shown in Fig. Tn7.1C). The four transposition-related genes occurred in the order p85-a, p54, p40 and p85-b from the left end (Fig. Tn7.1C) and, in view of the quality of the gel, could correspond to TnsB (80.8kD), TnsC (63kD), TnsD (59.1 kD) and TnsE (61.2 kD) respectively.
TnsA was presumably too small to detect in these experiments. There was some confusion as to the function of these proteins: while the authors observed Tn7 chromosome insertion not to require the p85-b (TnsE) and p40 (TnsD) products, as subsequently confirmed both genetically and biochemically (see e.g. [31]), plasmid insertion seemed to require p85-a (TnsB), p85-b (TnsE), p54 (TnsC) but also p40 (TnsD).
However, a more resolutive set of studies used genetic complementation of a plasmid-carried chloramphenicol-resistant mini-Tn7, with intact ends but no Tn7 genes, and compatible expression vectors with cloned Tn7 DNA fragments [31]. These identified five gene products required for Tn7 transposition: four (TnsA, TnsB, TnsC and TnsD) were essential for attTn7 site-insertion and the fifth (TnsE) was required for plasmid insertion whereas TnsD was not. Furthermore, two promoters (P1 and P2) located at the Tn7R were identified (Fig. Tn7.1B; Fig. Tn7.1Civ) using transcriptional and translational fusions and suggested that tnsA, tnsB and tnsC are translated in the same direction. P1 was also identified using S1 mapping. It was noted that expression from P1 was repressed by TnsB [31].
Overall, these results are globally consistent and identify five proteins involved in Tn7 transposition and two asymmetric ends each with multiple repeated sequences [26][27][28][29][30][31][32][33]. Moreover, they suggested that Tn7 uses two major transposition pathways, insertion into attTn7 and insertion into plasmids, and that these involve an overlapping set of Tns genes (TnsAB and C) and a specific Tns gene TnsD (attTn7 insertion) and TnsE (plasmid insertion).
Tn7 Function
The Major Assay Systems Used to Analyse Tn7 behavior in vivo
Analysis of Tn7 transposition has used three major in vivo genetic systems: mating out, λ hop and papillation assays.
Mating-out Assay
This assay [34] uses the conjugative plasmid pOX38 [35] as a target. This is composed of the large HindIII fragment of the F plasmid generally with a cloned drug resistance gene. For Tn7 studies, the resistance gene of choice was gentamycin (Gm), pOXgen [36]. pOXgen can be designed to carry a cloned empty attTn7 or an occupied attTn7. The transposon donor is either the host chromosome or a Tn-carrying plasmid. Various transposition functions are provided either from the chromosome or from compatible plasmids in the same strain. Mating into a suitable recipient strain and selection for the conjugative plasmid (Gm) and a suitable transposon marker gives a measure of the transposition frequency.
λ hop Assay
This assay uses an integration- and replication- defective derivative of bacteriophage λ [37] which can be propagated in a permissive host strain but which is defective when infecting a non-permissive host. Here, the fate of a phage- associated miniTn7 element carrying a kanamycin resistance gene is followed after infection of a non-permissive host.Inheritance of stable kanamycin resistance is a measure of transposition. Transposition functions are supplied either from the host chromosome or from plasmids in the infected strain.
Papillation assay
This assay, first developed for Tn10 [38] and Tn50 [39], involves a miniTn7 with no transposition functions but carrying a promoterless β-galactosidase gene and located in a transcriptionally silent region of a plasmid. Transposition into actively transcribed genes gives rise to lac+ papillae in a lac- background. Various transposition functions are provided either from the host chromosome or from compatible plasmids in the same strain.
Genetic analysis of TnsA-E Gene Expression and Function
The idea that Tn7 possesses different pathways for insertion (one into its unique att site and a second into plasmid targets) was suggested from early studies which initially identified Tn7-specific proteins [30] and was reinforced in complementation studies by the observation that TnsE was necessary for Tn7 insertion into a plasmid without a cloned attTn7 site [31].
Defining the Tn7 Open Reading Frames
These requirements were further refined genetically [40] using a three-plasmid system in a mating out assay [34]: a transposon donor carrying a minimal Tn7 devoid of transposition genes, pOX38 with and without a cloned attTn7, and a plasmid carrying cloned Tn7 transposition genes which had undergone insertional mutagenesis using a mini-MuΩ , a minimal derivative of the transposable bacteriophage Mu [40]. Ω is a strongly polar element [41]. The insertions were used to define the length of each Tn7 transposition gene, their genetic organisation and their effect on transposition into pOX38 or pOX38::attTn7 where it was found that tnsA, B and C were required for transposition activity and tnsD and tnsE were required in determining insertion into pOX38::attTn7 or pOX38 respectively.
Mini-MuΩ insertions into tnsA were polar on tnsB indicating that the two genes are probably part of the same transcription unit [40][42]. Similar experiments suggested that tnsC did not form part of a tnsA-tnsB operon. Additionally, insertions into tnsC were not polar on tnsD and tnsD::mini-MuΩ mutants were unaltered in tnsE-dependent transposition indicating that tnsD and tnsE are not part of the same operon. The authors also concluded that tnsC and tnsD form separate transcription units [40].
The DNA sequence of the tnsA-E region [43] had revealed five significant compact open reading frames (ORFs) (see also [42]): TnsA (273 amino acids, 31 kd); TnsB (702 amino acids, 81 kd); TnsC (555 amino acids, 63 kd); TnsD (508 amino acids, 59 kd); and TnsE (538 amino acids [29], 61 kd). These are all oriented in the same direction from Tn7R (Fig. Tn7.1A) all with potential initiation codons very close to the preceding termination codon: for tnsA and tnsB, the orfs overlap slightly (ATGTGGCAAATTAATGA); for tnsB and tnsC initiation and termination codons overlap (ATGA); for tnsC and tnsD the termination and initiation codons are separated by an intervening two base pairs (TAGCCATG); and for tnsD and tnsE the termination and initiation codons are adjacent (TGAGTG). These arrangements suggest that translational coupling [44][45] may occur [42].
Although the amount of TnsC observed when tnsB is disrupted by a polar insertion mutation [42] was considerably reduced, confirming that the tnsAB promoter and/or TnsB probably play a role in tnsC expression, some TnsC was still detectable. This was consistent with the idea [40] that, in addition to transcription from the tnsA-B promoter, tnsC can be transcribed from a second (tnsC) promoter. TnsC was observed to include a nucleotide binding motif [43] (In vitro transposition using purified Tns Proteins).
AttTn7, the Role of TnsD and Damage Control
AttTn7: a Safe Haven
Using a bacteriophage λ hop Assay , a small 68-bp DNA fragment including the specific Tn7 insertion point and extending upstream into the 3’ end of the glmS gene for about 13 codons was found to be sufficient for site- and orientation-specific Tn7 insertion [46] (Fig. Tn7.1Biii). A similar result was also obtained using a different assay system based on bacteriophage M13 transduction [47] where it was concluded that attTn7 is contained within a 64 base-pair region and sequences adjacent to the actual insertion site and encoding the 3’ end of the glmS gene were required. Thus, Tn7 must recognise the 3’ end of the glmS gene, which is highly conserved among bacteria, and inserts “safely” into a non-coding region located downstream without “damaging” glmS. Indeed, experiments in which mutations at the point of Tn7 insertion in attTn7 were introduced (by selection for glmS-ter mutations) showed that the sequence of the insertion point was not important for insertion [48].
TnsD Binds to attTn7
TnsD was shown to bind attTn7 since a cell extract from a strain carrying a TnsD-expressing plasmid formed a specific complex with a DNA fragment carrying the attTn7 sequence [49]. Moreover, a DNA fragment comprising nucleotides +55 to +28, while showing reduced insertion activity, still conferred insertion specificity. This sequence included a short, perfect inverted repeat (Fig. Tn7.1Biii). It was proposed that TnsD is likely to provide the sequence-specificity to the Tn7 transposition complex and may recognize the inverted repeat.
Although Tn7 inserts principally into attTn7 in the chromosome, four additional chromosome insertion sites which are used only rarely have been documented [50]. To achieve this, the authors used a host strain already carrying an occupied attTn7 which prevents insertion of a second Tn7 at this site (see Immunity) [12][33][47]. The secondary sites show significant identity to attTn7 particularly in the +25 to +62 region (Fig. Tn7.1B) which is required for site recognition.
In vitro transposition
A significant advance in understanding Tn7 transposition was the development of a cell-free Tn7 transposition reaction [51] since it opened the possibility of detailed biochemical analysis.
In vitro transposition using Tns cell extracts
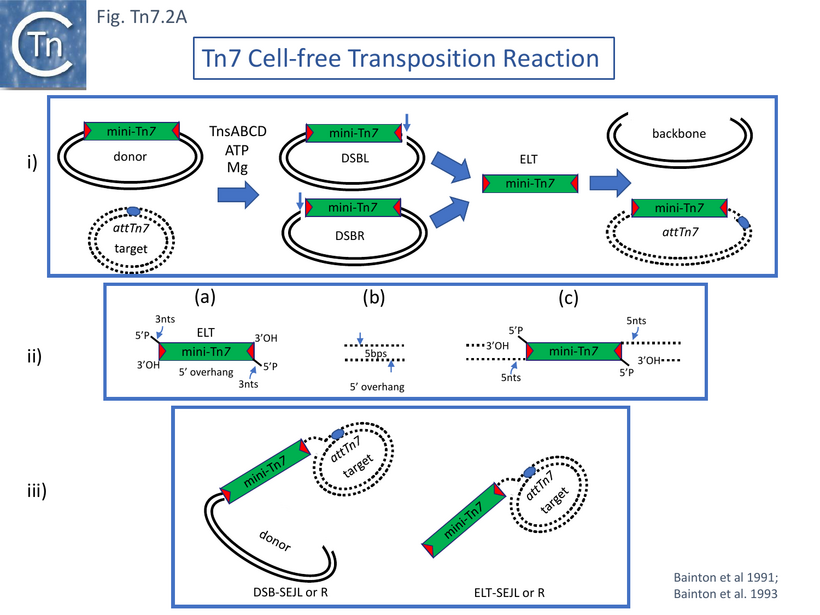
Incubating donor mini-Tn7-carrying and target attTn7-carrying plasmid DNA with fractionated cell extracts, ATP and, after a preincubation period, magnesium acetate, revealed a number of new DNA species: donor plasmids with double strand breaks (DSB) at Tn7R or Tn7L (DSBR and DSBL); species with the size of an excised mini-Tn7 and the equivalent linear gapped donor plasmid backbone; and species in which the mini-Tn7 had integrated into the attTn7-carrying target (Fig. Tn7.2Ai). The excised mini-Tn7 (ELT; Excised Linear Transposon) and the DSBs were found to be generated by staggered cleavage with 5’ overhangs (Fig. Tn7.2Aiia). The insertion products, as expected, occurred in the same orientation and, in the vast majority, at one of two adjacent nucleotide positions in attTn7. Moreover, the reaction also functioned when four partially fractionated extracts obtained from strains each containing a plasmid expressing a single TnsA,B,C or D gene product, were mixed.
Transposition did not require replication indicating that Tn7 transposes by a “cut-and-paste” mechanism. Moreover, high levels of Tn7 transposition stimulated recombination at the Tn7 locus in vivo implying that transposition creates a double strand break by Tn7 excision from the donor molecule [52] and that this acts as a substrate for recombination.
The reaction absolutely required ATP, extracts of all four proteins and both DNA substrates: donor and the target attTn7 [51]. Thus initiation of transposition chemistry necessitates the recruitment of target DNA into the transposition complex or transpososome.
In addition, an “excised” mini-Tn7 whose ends had been engineered to contain a restriction site with a 5’ 4bp overhang and a 3’OH at the Tn7 end was generated by digestion. This linear excised transposon, resembling an ELT, was able to undergo site- and orientation-specific insertion into attTn7 in this reaction suggesting that it could be an intermediate in transposition.
The notion was supported by the kinetics of the appearance and disappearance of the ELT: it appeared early in the reaction and decreased in abundance as the reaction progressed. The ends of the mini-Tn7 excised in the cell free reaction had a 3 nt 5’ overhang (Fig. Tn7.2Aiia) and insertion into the target, which would also have a 5’ nt overhang (Fig. Tn7.2Aiib), generated a gapped molecule (Fig. Tn7.2Aiic) which, when repaired, would generate the expected 5bp target repeat. The authors suggested that transposition occurred using a multipart intricate nucleoprotein complex and, furthermore they speculated that TnsA and or TnsC act as linker proteins assuring correct juxtaposition of TnsB-bound transposon ends and TnsD-bound target guaranteeing fruitful interaction at the Tn7 termini and attTn7 strand cleavage positions.
In vitro transposition using purified Tns Proteins
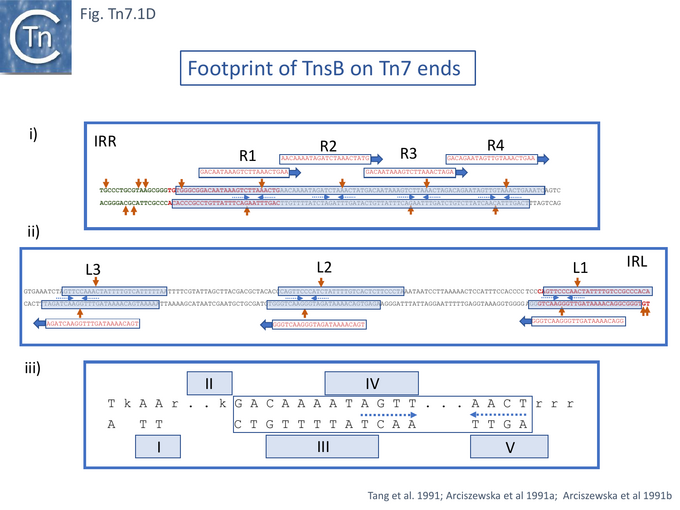
The cell free system was refined by using purified TnsB [53], TnsC [54], TnsA and TnsD [55]. For TnsB, the purified protein had been shown to bind the 22bp asymmetric repeats located at Tn7R and Tn7L[53] (Fig. Tn7.1D).
It had been noted from its sequence that TnsC carries a nucleotide binding motif [43] (see Fig. Tn7.3F and G). This was confirmed in vitro following purification of the TnsC protein [54]. TnsC was essential for transposition in a cell-free system [51] and bound adenine nucleotides (see Gain of function mutants, Nucleotide binding and hydrolysis). It was therefore probable that the protein is involved in binding the ATP co-factor required in the in vitro transposition system and indeed TnsC bound DNA in a non-sequence-specific way in the presence of ATP or of the non-hydrolysable ATP analogues AMP-PNP and ATP-γ-S but not in the presence of ADP [54].
As in the case of the crude extracts [51], the reaction with purified protein required: a donor plasmid with a mini-Tn7, an attTn7-carrying target plasmid, ATP and MgAc [55]. It was highly efficient and revealed not only the donor molecules with a single DSBL or DSBR and the ELT (Fig. Tn7.2Ai) but DNA species with the exposed Tn7L end of DSBL joined to a single attTn7 target DNA strand (DSB-SEJL and DSB-SEJR) or the ELT joined to a single DNA strand (ELT-SEJL and ELT-SEJR) (Fig. Tn7.2Aiii). The latter was present at very low levels.
Moreover, non-sequence-specific insertion was found to occur efficiently with TnsA, TnsB and TnsC alone if AMP-PNP was substituted for ATP, confirming that it is TnsD which directs transposition to attTn7.
A Model for TnsC/D attTn7 Interactions.
This study [55] also demonstrated that the attTn7 segment +28 to +55, which showed some target activity in vivo, was less active than the full attTn7 [49] and bound TnsD less well in gel shift assays thus providing a correlation between TnsD binding and insertion activity.
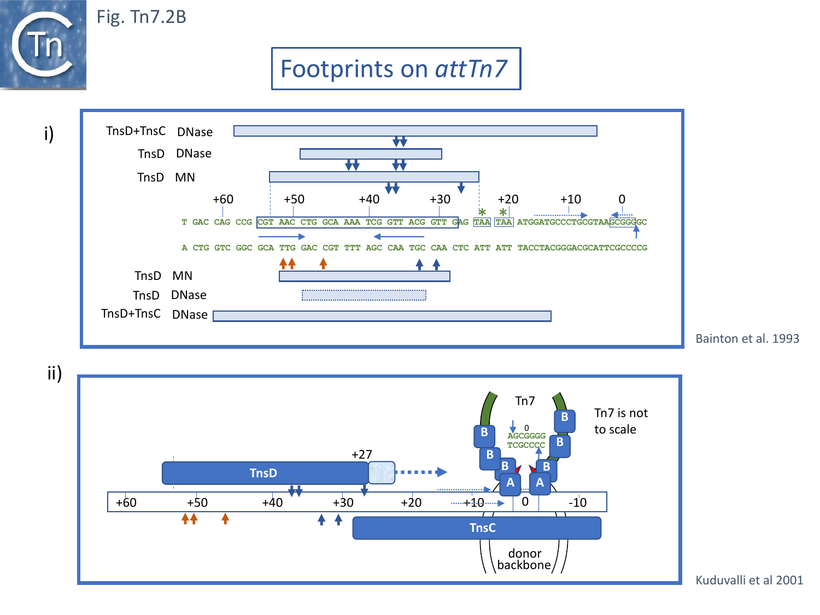
Gel shift assays had revealed that TnsC and TnsD form a distinct complex on attTn7 requiring ATP and disappearing in the presence of MgAc [54]. That this is a specific interaction which does not involve the non-specific TnsC DNA binding properties was evidenced by its stability in the presence of excess non-specific DNA [55]. Footprinting with micrococcal nuclease and DNaseI indicated that TnsC extended the TnsD protected region towards the Tn7 insertion point although this itself is not protected and therefore does not stably interact with TnsC or TnsD. (Fig. Tn7.2Bi).
Further analysis using more precise techniques such as hydroxy radical footprinting and missing base analysis to investigate TnsD-attTn7 interactions [56] indicated that TnsD not only contacts the core attTn7 region from +30 to +55 (Fig. Tn7.2B) largely in the major groove, but also recognises the +28 to +30 region distorting it towards the insertion site. The authors suggest that TnsC may recognise this distortion to bind and extend the TnsD footprint. They further suggest that the insertion site itself may be occupied by TnsC in the minor groove leaving access for transposase in the major groove, which is compatible with an attack by Tn7 3’OH ends across the major groove to generate a 5bp target repeat (Fig. Tn7.2Bii).
Triplex DNA as a Target
Interestingly, it was shown that triplex DNA also acts as a lure for Tn7 [57][58] in a Tn7 TnsABC in vitro reaction using a TnsC gain of function mutant, TnsC(A225V) no longer requiring TnsD. The DNA triplex formation can also induce neighboring conformational changes [57] resulting in insertions as for TnsABC+D insertions, at about 27-28bp from the DNA distortion [57][58] suggesting that the triplex-induced DNA distortion mimics that of TnsD-induced distortion and that this may be a recognition signal for TnsC.
Host Factors in Tn7 Transposition
It had been observed that crude cell extracts stimulated TnsD binding to attTn7 in the purified system and resulted in a lower mobility TnsD-attTn7 complex, host-TnsD-attTn7, in gel shift assays. These extracts did not bind attTn7 alone [55]. Further fractionation of the extracts revealed that the activity was due to two host proteins: acyl binding protein (ACP) and ribosomal protein L29 [59]. ACP stimulated formation of L29-TnsD–attTn7 complexes. Interestingly, ACP had previously been suggested to stimulate 3’ end-cleavage of the Tn3 transposon [60]. ACP exists in two forms: as a monomer and as a covalently linked dimer joined via disulfide bonds [61][62]. Both forms interacted with L29-TnsD–attTn7: the monomer generated only one defined band. The dimeric form generated a number of defined bands but, after reduction with DTT, these were reduced to the single monomeric band. Both L29 and ACP were physically present in the host-TnsD-attTn7 complex and stimulate transposition in the purified in vitro assay under limiting TnsD conditions [59].
Strand Transfer Reactions of Tn7 Ends.
The end-cleavage and strand transfer reactions were explored in vitro the TnsABC+E system and substrates carrying various mutations within their ends [63].
Strand transfer into the attTn7 target was addressed using an ELT (Fig. Tn7.2Aiia) produced by cleavage of appropriately positioned engineered restriction sites which generate flush 3’ OH ends and 4nt 5’ overhangs and which was found to undergo efficient insertion in the cell-free system (see above) [51]. A simple insertion (Fig. Tn7.2Aiic) was found to be the major product generated in a purified in vitro system although significant levels of ELT-SEJL and ELT-SEJR (Fig. Tn7.2Aiii) were observed together with products representing two R ends from two different ELT molecules joined to a single attTn7 (one to the top and one to the bottom strand). These might be expected as it had been shown that Tn7 derivatives formed from two Tn7R segments are competent for transposition [12].
In addition, if an equimolar mixture of separate (by restriction digest) Tn7R and Tn7L was used in the reaction, only Tn7R was found to integrate and this occurred in a concerted manner using two Tn7R although Tn7L was used efficiently if covalently linked to Tn7R. The strand-joining reaction required all 4 Tn7 Tns proteins: TnsA, TnsB, TnsC and TnsD.
Importance of the Teminal Dinucleotide
Like a number of transposons [64], Tn7 terminates with a CA-3’ end which is used to attack the target DNA. A mutant ELT terminating with GT-OH3’ (instead of CA-3’OH) at both ends was largely inactive in the insertion step. If only one end carried the mutation, the non-mutant end was transferred to the target but the mutant end was not. However, simple insertions could be detected with the single end mutants but at a very low level. Moreover, insertion of Tn7 mutants (with one -CA-3' end and one mutant -GT-3' end) occurred at the correct attTn7 position and in the correct orientation. This low level rescue of the mutant end is reminiscent of the genetic results obtained by Tang et al [65] as well as those obtained with other transposons including retroviruses [64][66][67][68].
No double-strand break could be detected at the target site in these experiments although both ELT- and DSB-SEJ species were efficiently generated. This led the authors to propose that Tn7 transposes by separate attacks of individual transposon ends on each target strand.
Donor Strand Cleavage Reactions of Tn7 Ends.
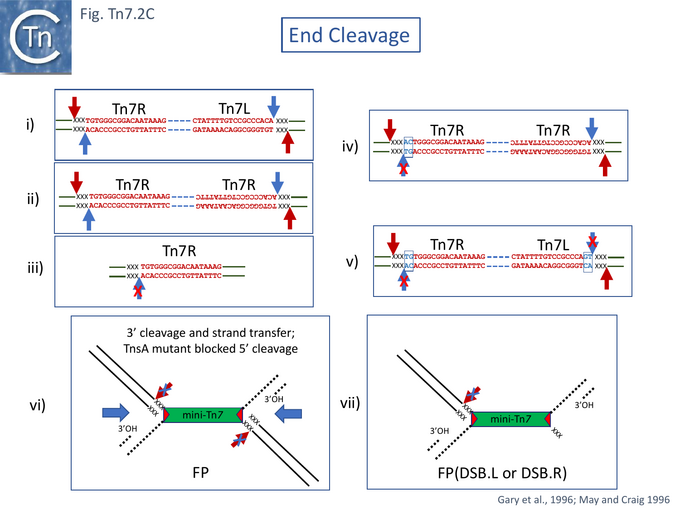
Although donor cleavage occurs on a donor plasmid substrate with two correctly oriented Tn7R ends in vivo [12] (Fig. Tn7.2Cii), no donor breakage could be observed with a plasmid substrate containing only a single Tn7R end (Fig. Tn7.2Ciii) indicating that double-strand breaks require the presence of two transposon ends on the same substrate DNA molecule [63]. Using supercoiled plasmid chimeric substrates with a -CA-3' end and a -GT-3' end, breakage and joining occurred normally at the -CA-3' end but not at the -GT-3' end (Fig. Tn7.2Civ). Thus, initiation of recombination requires two ends but the second end does not need to be active. However, while 3’ end cleavage was blocked on a -GT-3' end, substrates with two mutant ends were found to undergo 5’ end cleavage (Fig. Tn7.2Cv). This occurred 3nts outside (5’ to) the Tn7R and Tn7L both at wildtype and mutant ends, and its efficiency appeared to be influenced by the sequence of the flanking DNA. These data indicated that 3' and 5' end cleavage result from distinct activities.
The TnsA-TnsB transposase complex and a switch between cut-and-paste and Replicative Transposition.
Mutation of TnsA Results in Branched (Shapiro) Products in vitro
A number of studies have addressed the roles of and relationships between TnsA and TnsB. Using a mutant in TnsA (D114A) in the defined in vitro system, May and Craig [69] observed that instead of the expected reaction products (Fig. Tn7.2A), a novel species was produced. This proved to be a branched fusion molecule (FP) comprised of the donor and target replicons linked by a copy of the Tn7 derivative (Fig. Tn7.2Cvi) resembling the branched Shapiro intermediate [70] which occurs during transposition of Tn3 family members and bacteriophage Mu [70]. The TnsA mutation had blocked 5’ end cleavage but had not affected 3’ cleavage or strand transfer. Addition of a mixture of mutant and wildtype TnsA generated new species in which a both 5’ and 3’ end cleavages had occurred at one or the other end of the fused molecule but in which the 5’ cleavage at the other had remained blocked called FP(DSB.L or DSB.R) (Fig. Tn7.2Cvii). FP(DSB.L) and FP(DSB.R) were produced in equivalent amount. This suggests that at least two TnsA molecules are involved in end cleavages. However, the 3' reactions remained dependent on TnsA: there were no breakage and joining products in the absence of TnsA (even a catalytically inactive TnsA) in the reaction. The results therefore suggest that TnsB is responsible for 3’ end-cleavage.
Mutation of TnsA Results in Cointegrates in vivo
The cointegration reaction was also reproduced in vivo using a mating out assay with the conjugative F plasmid derivative pOX38 carrying a cloned attTn7 as a target. It was estimated that >80% of transition products were cointegrates when TnsA (D114A) was supplied but these were undetectable with wildtype TnsA [69]. Whether replication of the transposon occurs from the 3’OH generated from the insertion event (Fig. Tn7.2Cvi) or from normal plasmid replication forks has not been determined.
TnsA and TnsB Form a Heteromeric Transposase
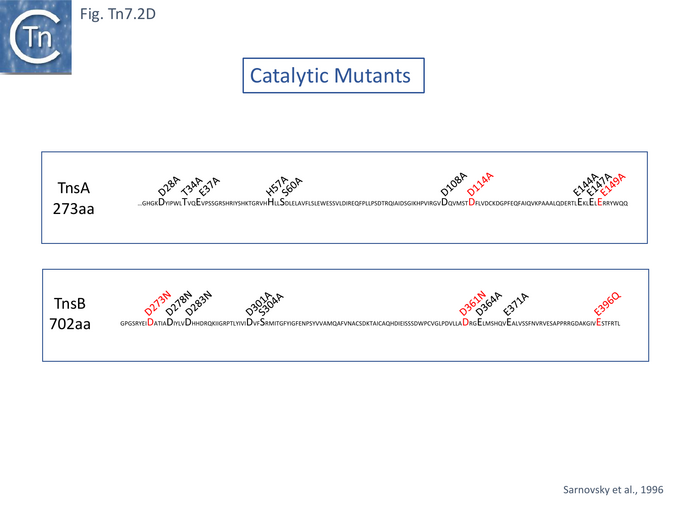
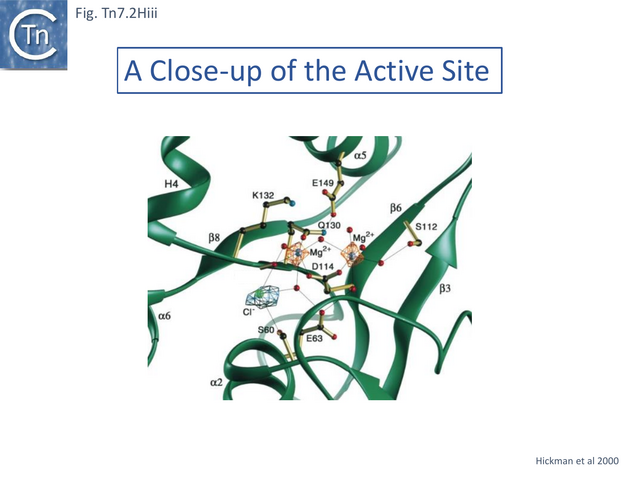
Further work from the same group established that the Tn7 transposase is heteromeric, comprised of both TnsA and TnsB which collaborate to generate the double strand breaks at each transposon end. This was assessed by using Tns mutants. It was proposed that both TnsA and TnsB carry a catalytic DD(35)E triad of amino acids typical of many transposases (Groups with DDE Transposases). TnsA possesses a poorly recognisable DD(35)E (see TnsA structure) while that in TnsB is more conserved. A number of potential DDE catalytic site amino acids were chosen in both TnsA and TnsB (Fig. Tn7.2D).
TnsA D114 which, when mutated, blocked 5’ cleavage in vivo and in vitro, appeared to form part of the proposed TnsA triad (Fig. Tn7.2D). Although no second, upstream D could be identified, mutation of the downstream E149 had a significant effect: in vitro, it resulted in a reduction (but not elimination) of 5’ end cleavage and also an increase in species at which one Tn7 end had a double strand break FP(DSB.L or DSB.R) (Fig. Tn7.2Cvii) with a preponderance of FP(DSB.L). It also generated simple insertions and DSB donor DNA molecules (Fig. Tn7.2A). This in vitro activity may reflect the inefficient recruitment of one of the neighboring E residues (Fig. Tn7.2D). TnsA therefore seemed to be involved in the functional difference between Tn7L and Tn7R. In vivo TnsA(E149A) behaved in a similar way to TnsA D114A.
Effect of the divalent metal ion on cleavage activity
The DDE triad is known to coordinate a divalent metal ion required for catalysis (Reaction mechanisms). Substitution of Mg2+ by Mn2+ has been shown to decease sequence specificity of a number of restriction enzyme [71][72][73] and to suppress the effect of mutations in the bacteriophage Mu transposase [74]. Like most transposition reactions, Tn7 transposition in vitro requires Mg2+. However, although the mutation TnsA(D114C) suppressed 5’ cleavage even in the presence of Mg2+, activity could be at least partially re-established if Mn2+ was used instead showing that TnsA D114 is involved in catalysis of 5’ cleavage.
Mutation of the potential TnsB DDE triad (Fig. Tn7.2D) also resulted in a recombination defect. In these cases, 3’ end-cleavage was affected while 5’ end cleavage occurred normally with added Mg2+. Moreover, Mn2+ was found to efficiently restore the 3’ joining reactions of D273N, E396Q and of D361N suggesting that these residues form part of the catalytic site.
The results reinforce the notion that TnsB carries a transposase-like DDE triad but are more ambiguous for TnsA (see below). They also suggest that TnsA not only executes 5’ end cleavage but also modulates 3’ cleavage by TnsB and, reciprocally, that TnsB is required for TnsA catalysed 5’ end cleavage possibly by positioning TnsA correctly at the Tn7 ends [75].
A Minimal Tn7 Recombination System Using Only TnsA and TnsB
To further emphasise that TnsA and TnsB together constitute the Tn7 transposase, a minimal Tn7 recombination system competent in both cleavage and strand transfer was developed [76]. This functions with wild type TnsA and TnsB, includes Mn2+ rather than Mg2+ and does not involve TnsC or require ATP. Breakage occurs predominantly at a single transposon end. The reaction proceeds by double strand breaks at either transposon end: Tn7L (DSB.L) or Tn7R (DSB.R) (Fig. Tn7.2Eii) with some intramolecular joining within the donor, principally to the other Tn7 end (Fig. Tn7.2Eii) to generate lasso structures (Fig. Tn7.2Eiii and iv) and Tn circles (Fig. Tn7.2Ev). Recombination required a supercoiled DNA substrate and both Tn7 ends. No cleavage occurred if either TnsA or TnsB was omitted from the reaction. The number of bases at the junction between 3’Tn7R (i.e. Tn7R as donor) and 5’Tn7L was observed to be either 3 or 4 (Fig. Tn7.2Eiii) whereas, for 3’Tn7L (i.e. Tn7L as donor) and 5’Tn7R, although the major species carried a 4 base spacer, a significant number of additional lengths were observed suggesting that the DSB at Tn7L and the position of joining was less precise than at Tn7R (Fig. Tn7.2Eii).
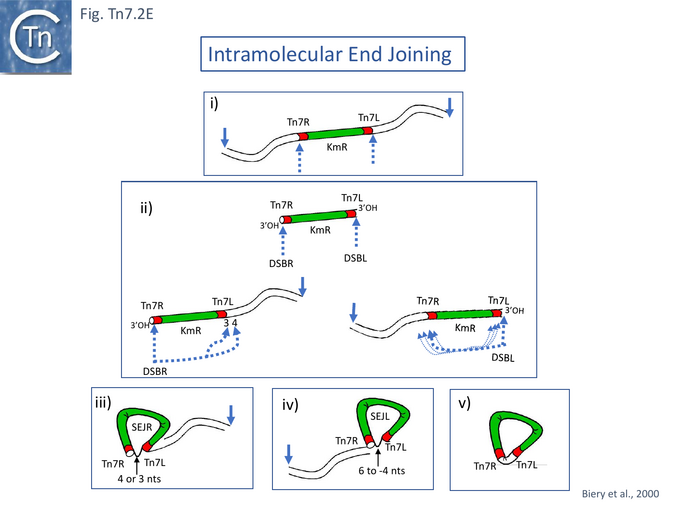
Functional analysis of Tn7 Ends
Some early attempts were made to analyse the sequence requirements at the very ends of Tn7R and Tn7L by mutagenesis [65]. In addition to the 22bp repeats, Tn7R and Tn7L include 8bp inverted terminal repeats similar to the organisation found in many transposable elements. The ends of many TE can often be divided into two functional domains (see: General Information/IS Organization): the terminal 3-4 base pairs which are required for the cleavage chemistry and more internal sequences recognised and bound by transposase. With a limited number of mutations in the terminal 8bp of Tn7R and Tn7L, using an M13 transduction assay, it was observed that the transversion of the penultimate G (G2->C)(Fig. Tn7.1D) at one or other Tn7 end resulted in a reduced transposition frequency to about 30% of wildtype but the same mutation at both ends resulted in undetectable transposition levels. Transversion of G6 (G6->C) had very little effect even when present at both ends. However, as might be expected, a more substantial set of mutations at the right end (TGTGGGCG -> ..AC.CGC) resulted in undetectable transposition levels but when these mutations were present at both ends, transposition was “rescued to a limited extent and increased to 1.5% of the wildtype.
Defining minimal Tn7 ends
Tn7 ends were further defined [12] using a mating out assay with pOX38 as target with or without a cloned attTn7: transposition was significantly higher into the attTn7-carrying plasmid than into the pOX38 vector when supplied with TnsABC+D; transposition into the pOX38 vector occurred when supplied with TnsABC+E. Using this approach, the Tn7 sequences required in cis for efficient transposition were narrowed down to 199 bp of Tn7R and 166 bp of Tn7L.
Further analysis showed that the initial miniTn7 donor composed of 199 bp of Tn7R and 166 bp of Tn7L could be further reduced to about 70 bp for Tn7R which includes the R1-3 22 bp repeats and 150 bp for Tn7L covering L1-3 (Fig. Tn7.1D). Whether transposition was measured with TnsABC+D (pOX38::attTn7) or with TnsABC+E (pOX38), the end deletions were found to have similar effects and are therefore presumably required in both processes.
Transposition in this system was inhibited in trans when high number of Tn7 ends were introduced on a separate plasmid (trans inhibition). Even copies of the R1 sequence alone produced some inhibition, suggesting that trans inhibition was due to competition for limited transposition proteins [12].
The analysis also demonstrated that while Tn7 derivatives constructed with Tn7L166 at both ends were unable to transpose using either TnsABC+D or TnsABC+E systems, those constructed with two Tn7R199 or Tn7R70 were active in both [12].
Tn7 Protein-Binding to Tn7 ends: TnsB and its Structure.
The complex ends of Tn7 are essential for transposition and several studies addressed Tns binding to the Tn7 ends [46][53][77][78][79]. Initially, it was observed that extracts from cells carrying Tn7 at the attTn7 site were “able to promote formation of discrete complexes with retarded (gel) mobility” with either radioactively labelled Tn7R or Tn7L ends [78]. These were eliminated when titrated with a large excess of the unlabelled opposite end indicating that they are end-specific. Cell extracts of strains without Tn7 did not generate complexes [78].
That the complexes involved only Tn7R or Tn7L sequences rather than their flanking attTn7 segments was demonstrated by the absence of complex titration (stability of the complexes) when an excess of unlabelled attTn7 sequences were added to the binding reaction. The same complexes were also generated by extracts of cells in which the tns genes had been cloned into a plasmid vector. Moreover, in this case, with higher basal Tns levels, several complexes could be detected, especially at higher extract concentrations, suggesting multiple binding sites. The extract also generated a single specific complex with a DNA fragment carrying only the L1 site. The binding activity was attributed to TnsB since extracts from cells with polar insertions obtained by insertion of a mini-Mu transposon into tnsC, D or E retained binding activity whereas a polar mutant in tnpB failed to generate complexes. Since there was evidence that the upstream tnsA and tnsB form an operon [40][42], an extract from cells carrying only the tnsB-E region was used and shown to generate identical complexes. Thus, TnpB alone was sufficient to bind both Tn7R and Tn7L.
Repression of Expression from the Transposase Promoter
As had been suggested [31], binding of TnsB “alone or in conjunction with tnsA” to the repeated sequences in the Tn7R end would explain the observation [31] that TnpB can repress expression from the transposase promoter since the -35 promoter component is located within direct repeat R4 (Fig. Tn7.1B). This was also supported by the observation that the low TnsA level observed from a plasmid carrying tnsA-E was increased when tnpB was disrupted [42].
Identification of TnsB Binding Sites
Using more purified TnsB preparations [53][77][79], addition of increasing protein concentrations resulted in the appearance of several bands on both Tn7R and Tn7L. Three complexes were observed for Tn7L, at lower protein concentrations, and as the protein concentration was increased, the lower migrating complex was chased into the middle and then the upper complex. With Tn7R, the same analysis revealed four complexes. These correspond to the number of 22bp repeated sequences in each end (Fig. Tn7.1D). Tn7R exhibited well defined more-or-less contiguous protection over the four repeats R1-4 in DNase footprinting experiments [53][77][79]. For Tn7L, the repeats L1, L2 and L3 were protected individually. In both cases, a number of hyper-sensitive DNase sites were observed suggesting that the bound DNA was distorted [53] (Fig. Tn7.1D). The terminal nucleotide at both Tn7R and Tn7L was unprotected and no TnsB-mediated DNA cleavage was observed even in the presence of additional Mg2+ (a divalent metal ion which had been shown to be important in related reactions e.g. [80][81][82][83][84].
The addition of the other partially purified Tns proteins had no effect on the protection pattern [77].
A more resolutive binding study [79] used hydroxy radical protection which probes close protein-DNA contacts revealed close small blocks of contacts alternating from one strand to the complementary strand along the length of each 22bp repeat (the consensus is shown in Fig. Tn7.1Diii). However, it was noted that for Tn7R, some of these were obscured due to the overlapping R1-R4 repeats. Moreover, the hyperactive DNase sites occur between the repeated elements of short palindrome (A G T T/C – A/G A C T) [79].
TnsB Structure in Complex with Tn7R DNA
A “tiled and intertwined” Structure
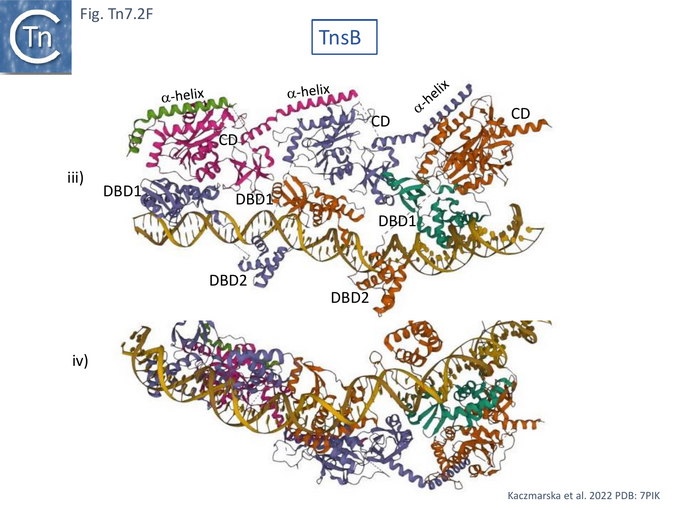
Detailed cryo-electron microscopy structures of TnsB bound to sites R1, R2 and R3 of Tn7R [85] revealed that the protomer folds into three domains separated by flexible linkers: an N-terminal DNA binding domain (DBD1) containing SH3 and helix-turn-helix subdomains; a second DNA binding domain, DBD2, composed of a winged helix (WH); and a catalytic domain, CD, with an RNase fold containing the D273, D361, and E396 active site triad [75] (Fig. Tn7.2Fi).
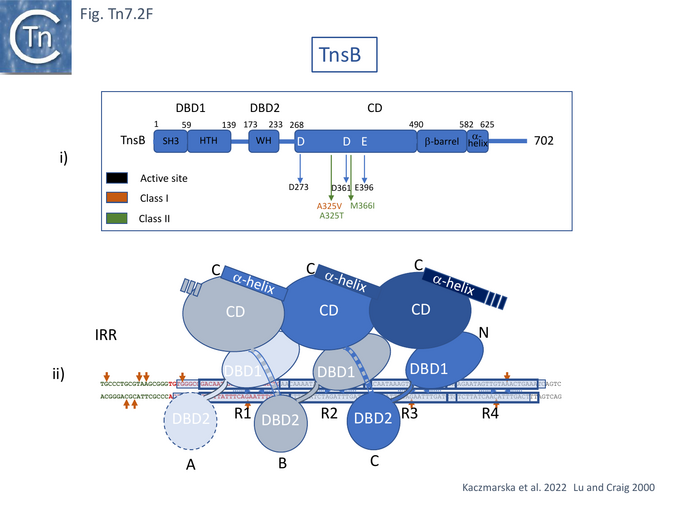
TnsB was observed to occupy three of the 4 R sites of Tn7R (Fig. Tn7. 2Fii, iii and iv) and introduce a significant bend in the DNA (Fig. Tn7. 2Fiv Fig. Tn7.2Gi and Gii) as previously inferred from mobility shift assays [79]. This model is consistent with previous footprinting data (Fig. Tn7.1D) [79]. It shows that sites R1, 2 and 3 are bound by the DBDs of protomers A, B, and C with DBD1 and DBD2 located only 3 bp from each other. The catalytic domains do not contact DNA but form extensive contacts with DBD1 such that the chain B CD interacts with chain A DBD1, chain C CD with chain B DBD1 and chain D CD with chain C DBD1 (Fig. Tn7.2Fii) in an intricately “tiled and intertwined” fashion [85]. The C-terminal α-helix also interacts with the preceding CD domain. Contacts are mostly with the DNA backbone. There are very few base-specific contacts and the authors show that Tn7R ends in which the sequences had been “scrambled” also bound TnsB indicating that binding shows sequence preferences rather than strict sequence-specificity.
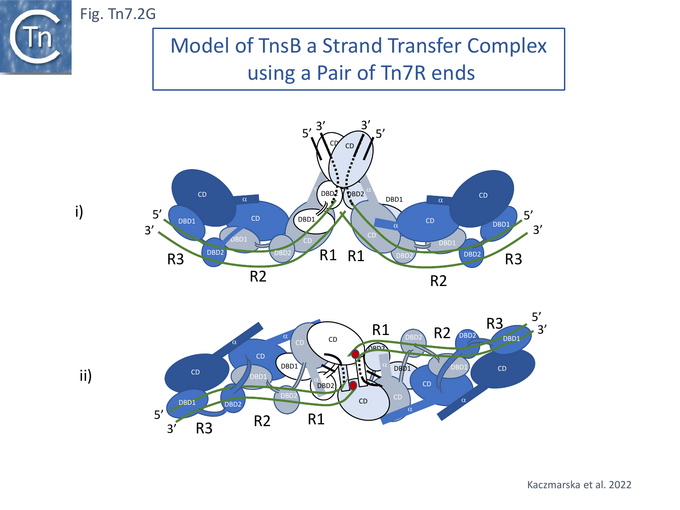
Modelling the Tn7 Paired End Complex (PEC)
TnsB exhibits similarities in structure to the phage Mu transposase, MuA, and this has led to the development of a model of the strand transfer complex (STC) based on the structure of the phage Mu STC [86] (Fig. Tn7.2Gi and Gii). Like Mu and several other transposons (Chemistry of the DDE enzymes), the catalytic TnsB subunit would carry out the reaction in trans, binding to one end while performing catalysis on the other. In the model, the chain A CD (not visible in the structure shown in Fig. Tn7.2Fii) must adopt a different conformation contacting both the transposon end and the target DNA. The trans configuration (Fig. Tn7.2Gii and iii) would explain the necessity for two Tn7 ends in order to catalyse transposition chemistry [63]. The STC as modelled, has no room to incorporate TnsA. This implies either that the model is incorrect (for example because it is built using two identical Tn7 ends or that TnsA is ejected from the transpososome complex before STC formation. However, the physical presence of TnsA but not its cleavage activity is required for TnsB activity [69].
It should be pointed out that the structural model uses a pair of Tn7R ends. Tn7R and Tn7L are organised differently with respect to TnsB binding sites and these differences are responsible for functional differences between the Tn7 ends: orientation- and site-specific insertion [9][33][46]; difference in length required for efficient transposition (about 150 base-pairs for Tn7L and about 70 base-pairs for Tn7R); and the fact that Tn7 derivatives formed from two Tn7R segments can transpose whereas elements formed from two Tn7L segments cannot [12].
TnsA Resembles a type IIS Restriction Endonuclease
Structural studies on TnsA [87] revealed the surprising fact that the protein has a close topological relationship with type II restriction endonucleases, specifically FokI, rather than a DDE transposase with an RNaseH fold (Fig. Tn7.2Hi). The active site was proposed to consist of E63, K132 and D114 (Fig. Tn7.2D; Fig. Tn7.2Hi and ii) [75] and included a pair of Mg2+ ions which were “liganded” to D114 (Fig. Tn7.2Hiii). This arrangement is similar to that observed in other endonucleases (see [87] for references). FokI is distinct from other type II enzymes since its catalytic domain cleaves DNA non-specifically at a fixed distance from is DNA recognition site (type IIS) and DNA sequence-specific binding is endowed by other domain(s) of the protein, three C-terminal HTH subdomains (Fig. Tn7.2Hi right) [88][89][90][91]. In TnsA, it was suggested that the helix in the C-terminal end is not involved in DNA interactions as it would be in the monomeric FokI but rather participates in protein-protein interactions with “another member of the transpososome [87]”.
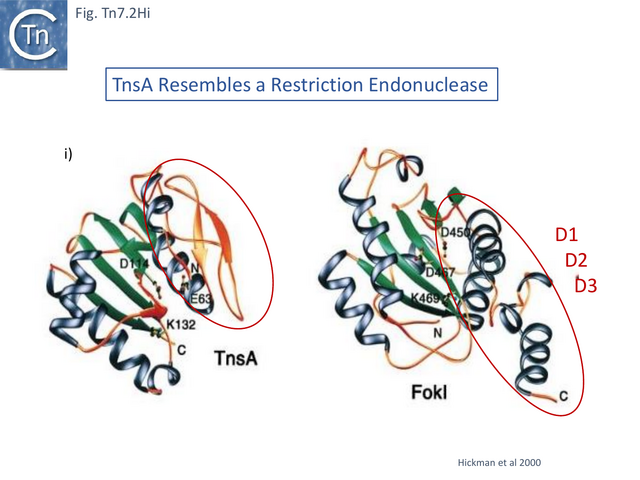
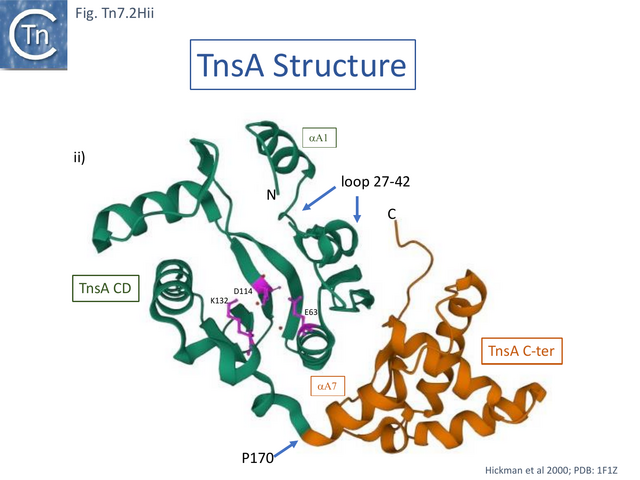
The TnsA structure could not be captured with bound DNA due to its low affinity, but binding was modelled based on other restriction enzymes [87]. It predicts that a loop following helix a 1 (residues 27–42) (Fig. Tn7.2Hii) would have minor groove contacts close to the 5’ cleavage site as is the case with some other restriction enzymes [87]while the N-terminal end of helix 5 would be involved in major groove contacts.
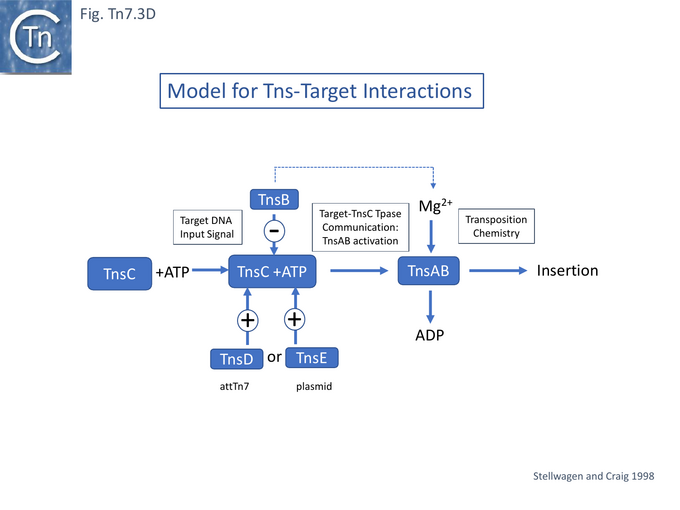
TnsA-B Gain of Function Mutants
A model elaborated for Tn7 transposition into attTn7 and conjugative plasmids [92] proposed that TnsAB activity is modulated by the ATP status of TnsC (TnsC Gain of function mutants, Nucleotide binding and hydrolysis) and that this is controlled by the TnsD-attTn7 target complex or by the TnsE-target complex which facilitate TnsC-ATP interaction. It was thought that the low level of insertion observed in the absence of TnsD-attTn7 is due to spontaneous TnsC-ATP formation [93].
Gain of Function Mutants with Increased Efficiency of Interaction with TnsC
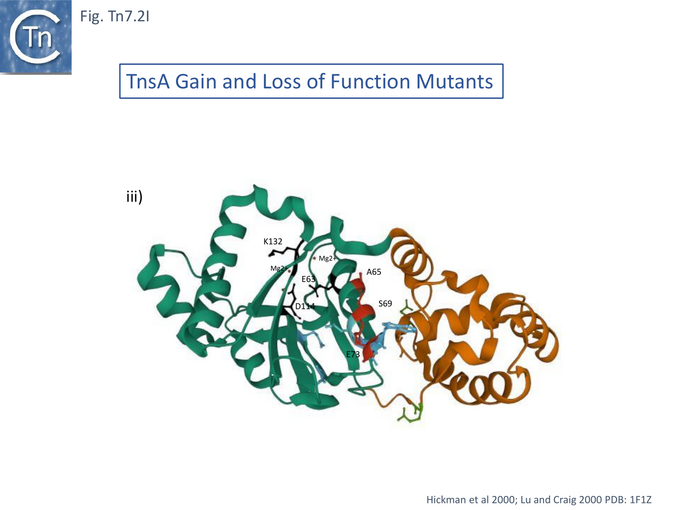
In studies aimed at probing TnsA-B function in the transpososome, the papillation assay was used to identify TnsA-B mutants which may exhibit increased efficiency of interaction with limiting amounts of TnsC-ATP (i.e. in the absence of TnsD-attTn7)[93]. A small number of TnsA (7) (Fig. Tn7.2Ii) and TnsB (3) (Fig. Tn7.2Fi) mutants were obtained showing increased activity in the presence of TnsC. These were divided into 3 classes all of which can be activated by TnsC alone: Class I mutants were activated even further by TnsC+TnsD or TnsE; Class II behaved like Class I but showed enhanced transposition activity in the absence of TnsC; and Class III did not undergo further activation by TnsD or TnsE. Combination of certain Class II TnsA and TnsB mutants permitted intermolecular insertion without TnsC.
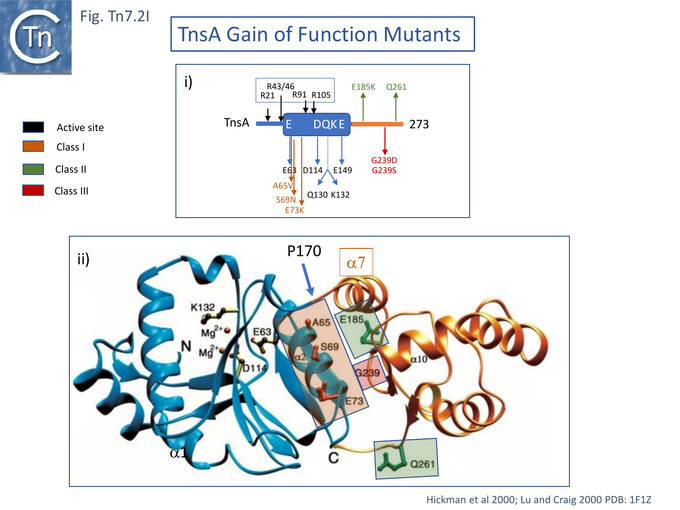
In the λ hop Assay , TnsC+D activated integration into chromosomal attTn7, for both Class I and Class II mutants while Class III mutant activity was too low for detection.
Effect of gain of function mutants in vitro
Examples of each class were purified and tested in the in vitro system: TnsA Class I S69N and Class II TnsA E185K were activated for strand cleavage by TnsC to generate DSD and ELT (Fig. Tn7.2A) as were Class II TnsB M366I and A325T. Activation was inhibited by ADP addition – suggesting that Class II mutants have enhanced interactions with TnsC-ATP. Combination of certain Class II TnsA and TnsB mutants in vitro without TnsC also generated DSB intermediates and circular transposon species.
Thus, class II TnsA and B mutants have intrinsically higher activities than wildtype and together are capable of supporting Tn7 recombination alone in the absence of TnsC confirming that TnsA+B constitute the Tn7 transposase.
Protease foot printing of TnsA
Protease footprinting [94] was used in an attempt to identify interactions between TnsA and TnsC. Alone, TnsA was observed to generate a number of cleavage products on partial trypsin digestion. Their endpoints at residues R21, R43/46, R91 and R105 are shown in (Fig. Tn7.2Ii), the R105 species proved resistant to further trypsin digestion suggesting that the TnsA tail is tightly folded. In the presence of TnsC, a large part of TnsA (R43/46, R91 and R105) became resistant to trypsin indicating that TnsC interaction probably results in a TnsA conformational change.
TnsC and Gain of Function Mutants
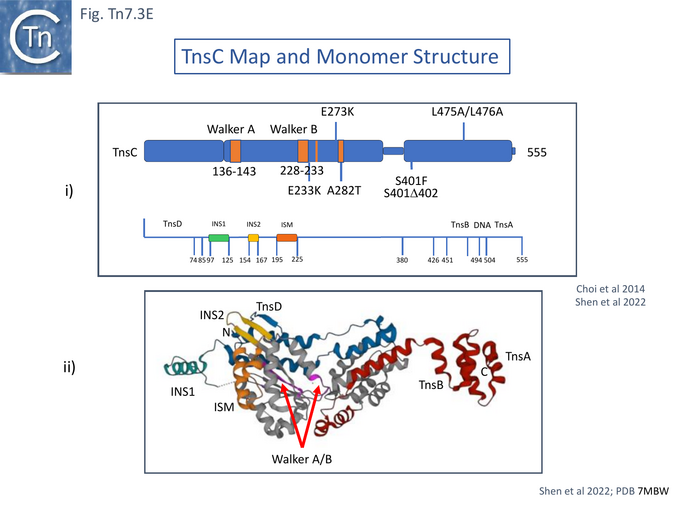
TnsC is a 555 amino acid long ATPase [54] with N-terminal Walker A and B motifs [43] (Fig. Tn7.3A) which interacts with TnsD and attTn7 DNA in an ATP-dependent manner [55]. It has been proposed to act as a connector (or molecular matchmaker [95]) between the TnsAB transposase and TnsD to engage the attTn7 target [55]. It was found to extend the TnsD footprint on attTn7 [55] (Fig. Tn7.2B).
Some low level Tn7 insertion had been observed in the absence of TnsD (i.e. with only TnsA, TnsB and TnsC) in the purified in vitro system and occurred into a number of different sites [55]. This low level TnsD-independent insertion was exploited as a means to screen for TnsC mutants permitting higher transposition levels [96]. Such gain-of-function mutants might be expected to activate transposition in the absence of TnsD (or TnsE).
Identification of TnsC Gain of Function Mutants
For this, a papillation assay [38][39][97] was used. With TnsABC alone, papillae were undetectable. With TnsABC+D only a few papillae were detected because of the absence of a suitable promoter in the glmS region whereas with TnsABC+E, a significant number of papillae occurred. The tnsC gene was subject to random mutagenesis and mutants were screened in a TnsABC assay to obtain 6 examples in which transposition was activated (Fig. Tn7.3A) and which gave rise to significant levels of papillae in the absence of TnsE (or TnsD).
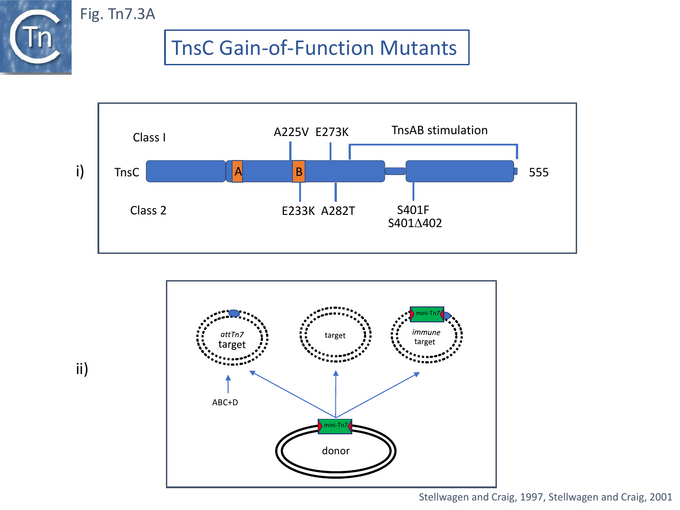
The mutants were of two types depending on their response to TnsD and TnsE: transposition activation by Class I mutants could be stimulated by TnsD and targeted to plasmid pOX38 by TnsE, while the activation by Class II mutants was not responsive to TnsD and/or TnsE stimulation.
For example, a λ hop Assay in which a miniTn7Kan carried by a defective phage is used to infect a recipient expressing various Tns genes including TnsC, showed that: TnsABC+D generated a large number of KmR chromosome insertions while those from TnsABC+E occurred 103 fold less frequently and TnsABC-mediated insertions were undetectable.
However, all TnsC mutants isolated in the papillation screen showed activity in the TnsABC configuration. The A225V and S401Δ402 mutants showed an order of magnitude activity increase compared to TnsABC+E while stimulation by the others was less marked.
Also, in a mating out assay, all showed activity. The most active was S401Δ402 but none were more active than TnsABC+E alone. Class 1 mutant A225V- and E273K-mediated attTn7 insertion was stimulated by several orders of magnitude whereas class 2 E233K, S401Δ402 and A282T were slightly inhibited.
TnsD-independent insertions (i.e. low level activity in the presence of only TnsA, B and C) occur at a low level with wildtype TnsC. However, using the TnsC A225V gain-of-function mutant, transposition frequencies approached those of TnsABC+D. An exploration of the target specificity of TnsABC insertion designed to determine the nature of the recognised target signal(s) revealed that the system preferentially directs insertion adjacent to triplex DNA [57]. It is known that triplex formation has a structural influence on the adjacent DNA (e.g. [98] and it was suggested that these distortions may provide a signal for TnsC recognition. This is compatible with the observation that TnsD binding distorts the DNA region towards the insertion site and the suggestion that TnsC recognises the distorted attTn7 region [56] (Fig. Tn7.2Bii).
Gain of function mutants, Nucleotide binding and hydrolysis
Purified TnsC was shown to bind ATP by UV crosslinking and to bind DNA non-specifically [54] and DNA binding was also reported to occur with dATP, ATP-γS and AMP-PNP. In contrast, no binding of DNA to TnsC was observed in the presence of ADP or AMP. Moreover, both ATP-γS and AMP-PNP supported better DNA binding than did ATP suggesting that ATP hydrolysis to ADP destabilises the complex. Indeed, this effect was exacerbated by addition of Mg2+ to the binding reaction while only a small reduction in binding occurred with ATP-γS. This ATP-dependent behaviour is also observed in the sequence-specific TnsD-facilitated TnsC binding to attTn7 where the complex (which is resistant to the addition of non-specific DNA and therefore not due to nonspecific DNA binding properties of TnsC)) disappears on addition of Mg2+ [54][55].
The in vitro TnsABC+D reaction absolutely required ATP, Mg2+, extracts of all four proteins and both DNA substrates: donor and target attTn7 [51].
TnsC A225V was found to be about 3 fold less active in ATP hydrolysis [99] than wildtype TnsC and it was speculated that this might contribute to the gain of function by allowing the mutant to spend more time in the activated ATP-bound state. It is also, like the wildtype TnsC, inhibited by ADP.
On the other hand, the class 2 mutants TnsC S401Y402 (Fig. Tn7.3A) showed similar levels of TnsABC transposition with ADP, ATP or the non-hydrolysable AMPPNP or indeed without any exogenous nucleotide at all [99] and it was proposed that these are locked into a “constitutive” conformation regardless of the nucleotide available.
Both TnsC S401Y402 and A225V mutants were also found to bind DNA more tightly being more resistant to increasing Mg2+ concentrations than wildtype TnsC and were retained more strongly on addition of competitor DNA.
These studies also revealed that TnsC can activate the TnsAB transposase for catalysis of donor cleavage in vitro. Using special reaction conditions (no target DNA or low Mg2+ incubation and high glycerol concentration) wildtype TnsC stimulated TnsAB-mediated donor cleavage in a concentration-dependent manner. This was mirrored (albeit to a 50 fold lower level) by a TnsC deletion derivative of the first 293 amino acids (Fig. Tn7.3A).
Thus nucleotide binding is required for formation of the TnsC-attTn7 DNA complex by presumably creating a favourable binding conformation (which, in gain of function mutants, is assumed without nucleotide binding) and TnsC stimulated the endonuclease activity of the TnsA/B transposase.
The TnsC C-Terminus Interacts with TnsA and TnsB
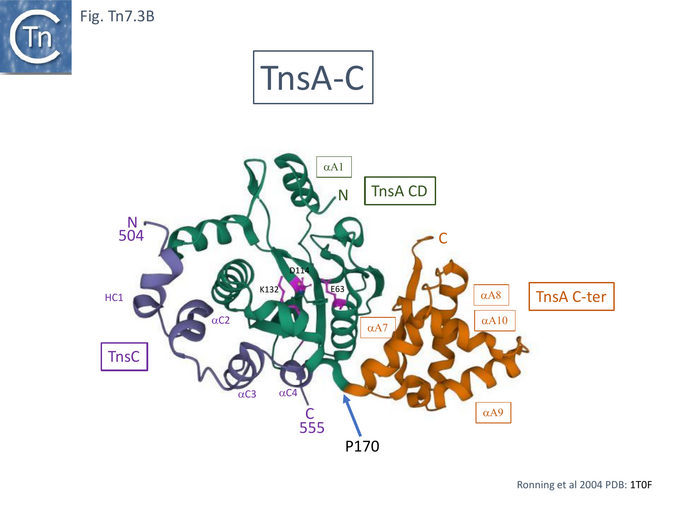
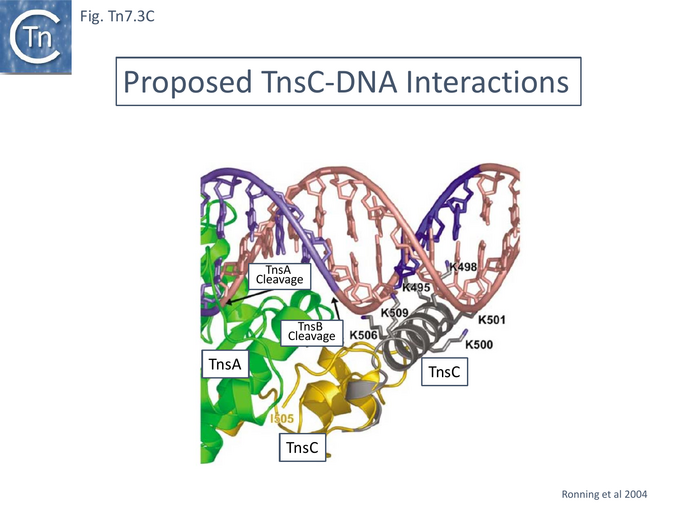
TnsA was shown, using affinity chromatography, to interact directly with both full length TnsC (1-555) and a C-terminal fragment (294-555) [99] and by protein footprinting where full length TnsC protects the N-terminal TnsA catalytic domain from partial protease digestion [93]. Coexpressed TnsA and TnsC form a tight complex and elute together from a gel filtration column with an apparent 2:2 migration time [100]. When the TnsA/TnsC complex was partially digested with papain, only a small TnsC fragment continued to bind TnsA. This proved to be TnsC (504–555). TnsA will not bind DNA alone, but the TnsA/TnsC 495–555 complex will bind DNA in as sequence-independent manner. These observations were extended by structural studies of TnsC (504-555) bound to TnsA [100] (Fig. Tn7.3B) where it was found that the interactions were exclusively with the catalytic domain of TnsA within a hydrophobic pocket and leave the catalytic residues available for the transposition chemistry. This is the region where Class I TnsA gain of function mutants map (Fig. Tn7.3A) [93] i.e those which allow TnsAB activation by TnsC in the absence of either TnsD or TnsE. Although binding of the TnsC fragment does not result in any gross structural changes in TnsA although TnsA alone includes two Mg2+ ions, in the TnsA/ TnsC (504-555) co-crystal (Fig. Tn7.3B) there is only one. However, TnsC (504-555) binding does not interfere with the proposed TnsA active site binding mode. Moreover, modelling TnsC to residue 494 (Fig. Tn7.3C) showed that a number of its surface exposed basic residues might in fact interact with DNA phosphates to enhance DNA binding to Tn7 ends [100]. Indeed, mutation of the basic residues within TnsC 494 – 504 abolished DNA binding by TnsA- TnsC 494 – 555 (Fig. Tn7.3E).
Stellwagen and Craig [92] had proposed a model in which TnsC is a central player in co-ordinating Tn7 transposition (Fig. Tn7.3D): it is activated by TnsD and recruited to the attTn7 target (whereas TnsE was proposed to play a similar role in recruitment to a conjugative plasmid target) but is inactivated by TnsB and removed from immune targets already carrying a Tn7 copy (see immunity below) and also contacts the Tn7 transposase TnsA/B to activate transposition chemistry. Indeed TnsB-facilitated dissociation of the attTn7-TnsC/D target complex was subsequently demonstrated directly [101][102] and shown to require only a C-terminal TnsB fragment: TnsB 677 – 694 [101]. This fragment was used to probe TnsB/TnsC interactions using a substituted with the photoactivatable crosslinker benzoylphenylalanine (BPA) [103] and Maltose Binding Protein–tagged fragments of TnsC and showed that the C-terminal region TnsC 451 – 555 (Fig. Tn7.3E) was sufficient for crosslinking to occur.
The TnsC 451-494 region contains a number of highly conserved residues among a set of 24 different TnsC examples [101]. Interactions TnsC/B interactions were further refined by examining the effect of mutating a number of these: in particular, L475A and L476A. When introduced into an MBP-TnsC 361-555 fragment, these mutations blocked TnsB crosslinking and considerable reduced or eliminated transposition in vitro and in vivo when introduced into full length TnsC. However, they did not block formation of the TnsC/TnsD-attTn7 target complex but did block both TnsB-facilitated disassembly of this complex and TnsC stimulated Tn7 end pairing [101]. Finally, these studies also showed that the N-terminal TnsC 1-85 region, could interact with TnsD 1-309.
TnsC Structure.
Use of a More Stable Shortened TnsC Derivative
The structural studies employed a shortened TnsC derivative since removal of the C-terminal region of TnsC, 504-555, which interacts with TnsA [100] (Fig.Tn7.3B), was found to improve protein stability and facilitate protein manipulation. Moreover, band shift analysis using a 15 bp duplex DNA indicated that TnsC could bind in a non-sequence-specific manner in the presence of ATP but gave a “sharper” band if ATP was substituted for the non-hydrolysable AMPPnP. This suggested that ATP hydrolysis destabilises the protein-DNA complex. Therefore, a TnsC derivative with lower ATPase activity, the A225V gain-of-function mutant (Fig. Tn7.3A) [99], was used. This derivative formed stable complexes with either nucleotide and exhibited nucleotide- and DNA-dependent oligomerisation, confirming that DNA binding requires only ATP binding and not hydrolysis (Gain of function mutants, Nucleotide binding and hydrolysis)[51][54].
A TnsC Protomer Crystal Structure
A crystal structure of TnsC was obtained showing it to assume an extended form [104] (Fig. Tn7.3Eii). The region 74-380 assumes a typical AAA+ fold with three insertions. One of these, residues 195–225 corresponds to the initiation specific motif (ISM) characteristic of AAA+ proteins from the initiator clade [105] which groups initiator/helicase-loader proteins, and, importantly includes the phage MuB and IS21 family IstB [105] transposition “helper” proteins. ISM is involved in DNA binding [104].
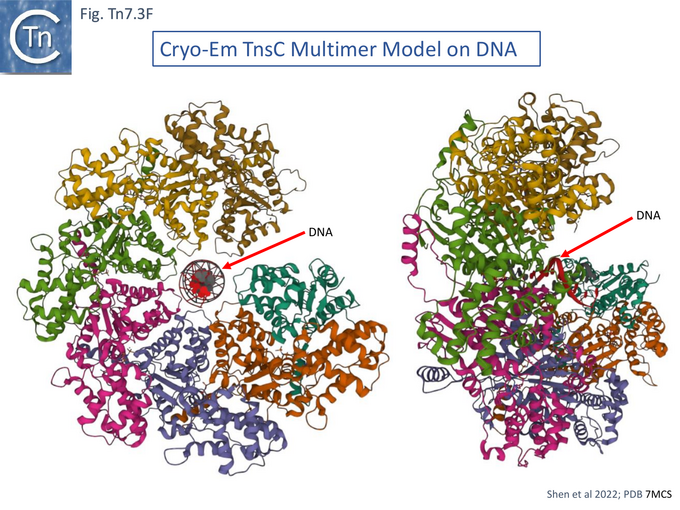
A Cryo Electron Microscopy Stucture of the TnsC-DNA Complex
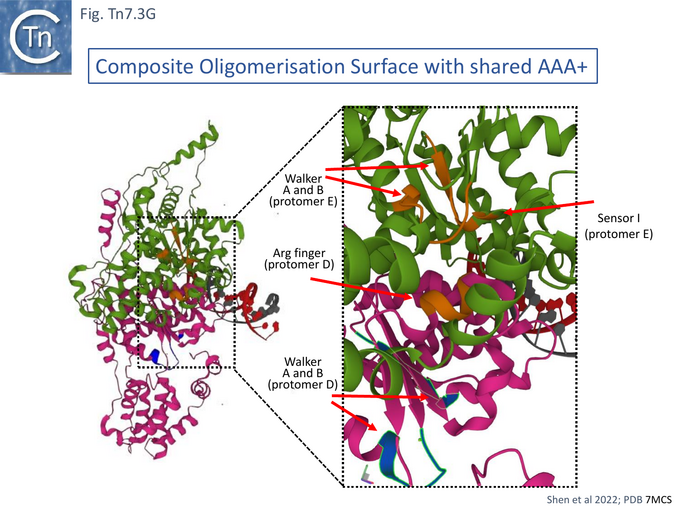
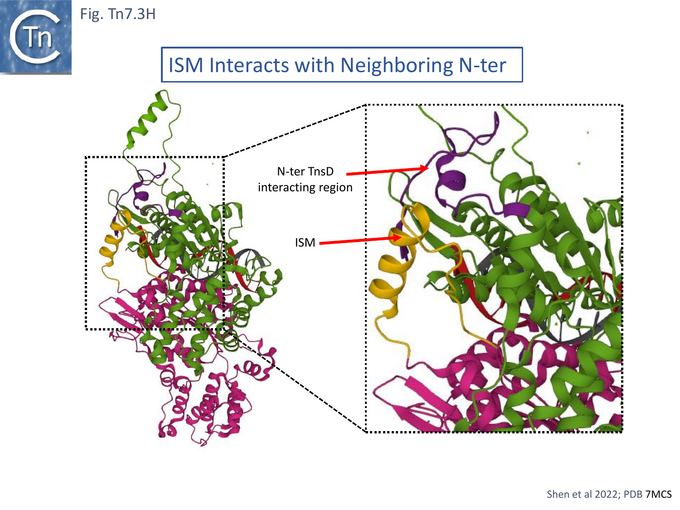
Cryo electron microscopy [104] also revealed that TnsC formed open, heptameric rings on DNA. Although TnsC can bind to 15 bp duplex DNA, 30 bp are necessary to accommodate 2 rings. A refined cryo-em map (Fig. Tn7.3F) revealed a heptameric cluster with a composite oligomerisation surface in which the Walker A (136-GCSGSGKY-143), Walker B (228-LIVIDE-233) and sensor I (T268) are provided by one protomer which a neighboring protomer provides the arginine finger (280-RSARR-284) (Fig. Tn7.3G) and in which the ISM interacts with the N-terminal end of a neigboring protomer (Fig. Tn7.3H).
The overall structure indicated that the N-terminal region of TnsC which interacts with TnsD and the C-terminal region which interacts with TnsA and B (Fig. Tn7. 3Eii) lie on opposite faces of the TnsC ring and the authors suggest that TnsC provides a partitioning function which coordinate, on the one hand, target interaction (TnsD), and on the other, paired end complex formation and cleavage (TnsA/B) [104]. The structure revealed that the N-terminal end interacts with the distorted region of DNA generated at attTn7 by TnsD (see Fig. Tn7.2B) [56]. The observation that TnsC does not make specific contacts with the distortion but interacts with neighboring duplex, they suggest, “provides a universal mechanism” to recruit TnsC to other types of target site (see TnsE and TniQ later).
A model based on the TnsC [104] and TnsA [56] structures (Fig. Tn7.3I) was developed which takes into account the observation that TnsD protects a segment neighboring the attTn7 distorted segment (Fig. Tn7.2B) and that TnsC binding extends this protection [55]. It proposes that by encircling DNA, TnsC determines the strict spacing between the point of insertion and the target site. Although the C-terminal TnsC region was missing in the cryo-em structure, the previously solved structure of TnsA bound to the TnsC C-terminal binding domain [100] (Fig. Tn7.3B) was used to extend the structure. It is represented as a “carousel” form in which TnsA docked into the flexible C-terminal ends and indicates the empty segment which would interact with TnsB (Fig. Tn7.3I). The authors propose that the bound TnsA molecules function as “beacons” which recruit TnsB bound to donor DNA, facilitating paired-end complex formation.
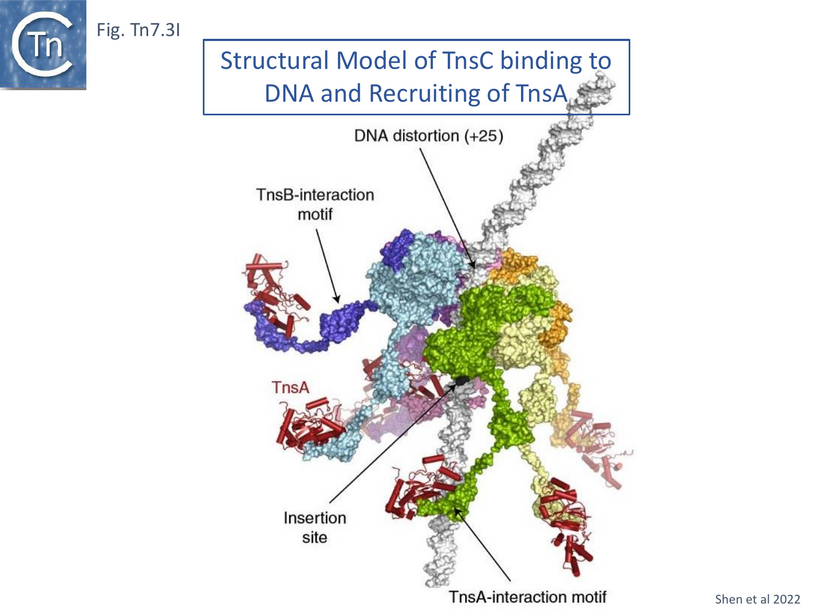
The model proposes that TnsC recruits TnsA and subsequently is itself recruited by target-bound TnsD (Fig. Tn7.3Ji) to form the TnsC ring (Fig. Tn7.3Jii and Jiii). The TnsA sequestered in the extended tail of the TnsC ring is then able, in turn, to recruit TnsB bound to Tn7 ends (Fig. Tn7.2F and Fig. Tn7.3Jiv) to generate the paired end complex (Fig. Tn7.3Jv and Fig. Tn7.2B). Indeed early experiments suggested that favorable attTn7 targets appeared to promote Tn7 excision from the chromosome and implies that the attTn7/TnsD/TnsC complex acts early in the transposition pathway [96].
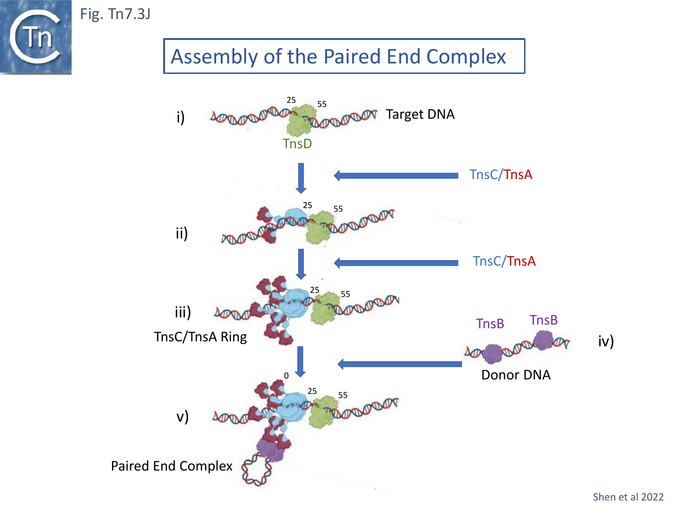
The role of TnsE
Together with TnsA, B and C, TnsE is required for insertion into plasmids which do not carry attTn7 [30][31]. It was observed that insertion sites into plasmids, tnsE sites, were very different from those involving TnsD [50].
Insertion Proximal to the E. coli Replication Terminus
Additionally, it was shown in vivo that, using the TnsABC+E gene set, Tn7 insertions show a strong regional preference for the terminus region of the bidirectionally replicating E. coli chromosome [106] where replication forks meet (Fig. Tn7.4Ai). The major regional hotspot occurred between the important Ter sites TerA and TerC which are closest to, and flank, the terminus. The Ter sites are sequences on the chromosome to which a protein, Tus, binds. They are oriented, and when bound by Tus, strongly reduce replication fork progression through Ter in one orientation (permissive) but not in the other (non-permissive).
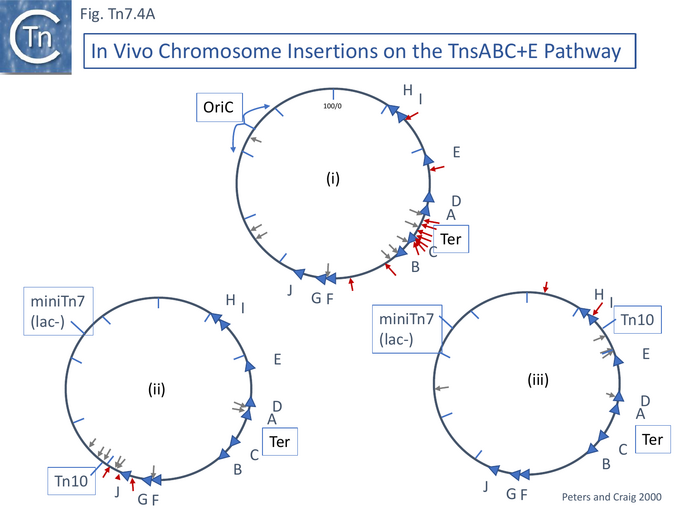
This system is designed to prevent replication forks escaping the terminus region and proceeding into the opposite half of the chromosome by interacting with the DnaB helicase (see [107][108][109][110][111][112]). It appeared the TnsE was necessary for the regional specificity since the pattern of insertion appeared random when a TnsE-independent mutation of TnsC (A225V) isolated as a gain-of-function mutant (Fig. Tn7.3A [99][113]) was used in the absence of tnsE (i.e. with only tnsABC). Moreover, ablation of tus did not eliminate the insertion preference although it did increase the extent of the target region. The authors suggested that in the tus mutant strain, Tn7 insertion continues to occur where replication termination takes place but that termination is no longer confined to the small chromosomal region as it is in the wildtype strain.
Targeting to Double Strand DNA Breaks
It was proposed that this targeting is due to the occurrence of double strand DNA breaks (DSB) in the region. It had been noted that homologous DNA recombination is highly stimulated in the ter region [114] and it was suggested that homologous recombination was stimulated by DSBs [115]. To investigate this, a system was designed in which induction of transposition of chromosomal copies of a derivative of the Tn10 transposon, capable of the excision step but unable to reinsert [116], was used to generate DSBs [117] together with a papillation assay. A stimulation in miniTn7 transposition of about 2 fold was observed for strains carrying the Tn10 insertion at two different chromosomal locations. This was dependent on both TnsE and on a functional (mutant) Tn10 transposase. Moreover, in both cases, insertions occurred in the region of the Tn10 location [106] (Fig. Tn7.4Aii and Fig. Tn7.4Aiiii) although in one of the Tn10 derivatives, the effect was mild (Fig. Tn7.4Aiiii) possibly because the sample size was small.
One explanation for the occurrence of some insertions at a distance from the break site (Fig. Tn7.4Aiiii) was that the DSBs are displaced by RecBCD exonuclease processing (which is also involved in the hyper-recombination in the terminus region [118]). This is influenced by the density of the polar, 8-bp ⬇ sequences which alter the RecA loading and exonuclease properties of RecBCD (see [119][120]). However, more detailed analysis [121] using three additional Tn10 insertions suggested there are small regional chromosome insertion hotspots (defined as multiple insertions occuring within a region of 1 kb of one another) which occur at fixed positions and are activated by neighboring DSBs. These, or the stimulation of Tn7 transposition, were shown not to depend on RecBCD and are therefore not dependent on the distribution of ⬇ sites. DNA damage caused by UV light, mitomycin C, or phleomycin also stimulated transposition but this is not affected by the SOS system or by genes involved in stalled replication fork restart.
One attractive idea is that the recognition signal for insertion is the recognition by TnsE of DNA replication structures involved in the repair of DNA double strand breaks [121][122]. It should be noted that another, unrelated transposon, Tn917, a synonym of Tn551 and a member of the Tn3 family, also shows similar affinity for insertion into the terminus region of the Bacillus subtilis chromosome [123]. Insertions are clustered around the two terminus proximal sites, terI and terII, and requires a functional RTP (equivalent to TUS) termination protein.
Insertion into Conjugating Plasmids
The observation that Tn7 shows a preference for insertion into plasmids which requires TnsE [30][31] and occurred in a specific orientation in each of the plasmids RP4. R91.5 and R388 [15][17][31] was further explored in more detail [124] using a λ hop Assay into strains supplying TnsABC+E and carrying different target plasmids: R388, pOXGen (a gentamycin resistant derivative of pOX38 [35]), the phage/plasmid P1 and the small high copy number plasmid pBR322. Insertion was found to occur almost exclusively into R388 or pOXGen and, in the case of the pOXGen-carrying strain, transposition was stimulated 50 fold. Neither the (much larger) host chromosome, P1 or pBR322 were found to be effective targets. Insertions into pOXGen occurred into many different sites but in only one orientation. The common feature of R388 and pOXGen are that both plasmids are conjugative. The orientation of Tn7 was such that the right end (proximal to the transposition genes; Fig. Tn7.1A) would be the first to enter the recipient cell.
The role of “conjugability” was confirmed by using a conjugation-deficient pOXGen derivative, pOXΔtraI containing a substitution of the traI gene (the relaxase) with a dhfr (dihydrofolate reductase; trimethoprim resistance) gene leaving the tra operon otherwise intact. The derivative plasmid became a poor Tn7 target with most insertions located in the chromosome. However, those insertions which are located in the plasmid occur in both orientations. When TraI is supplied in trans, pOXΔtraI, becomes an efficient target and insertions are oriented as they are in pOXGem.
Furthermore, a pBR322 derivative into which the functional 500 bp pOXGem minimal origin of transfer, oriT, became a proficient target when transfer functions were provided in trans from a resident F plasmid [125]. All insertions were oriented with respect to the direction of transfer. Supplying TraI in trans alone did not affect the target proficiency indicating that it is not the TraI-induced nick at oriT alone which is responsible but target proficiency involves additional transfer functions.
The question of where transposition into the conjugating plasmid occurs (in the donor strain in which rolling circle replication is occurring during transfer or in the recipient cell in which the complementary strand is synthesised; [126][127] see [128]) was addressed genetically [124]. The approach was to use a mating strategy using a donor carrying pOXΔtraI and a source of TraI but no transposition enzymes (Transfer+, Transposition-) and a recipient without a TraI source but with a transposon and a source of TnsABC+E (Transfer-, Transposition+). It was observed that Tn7 inserts preferentially into the transferred pOXΔtraI, implying that Tn7 targets the incoming plasmid transferred strand and may be associated with transfer replication.
A mechanism to promote transposon dispersion.
The TnsABC+E pathway is therefore designed to facilitate dispersion of the transposon. This dispersion is likely amplified since, in contrast to plasmid F and its derivatives which are depressed for their transfer functions and conjugate efficiently, transfer of many naturally occurring conjugative plasmids is controlled by repression and initially occurs at low frequency. However, when these enter a naïve cell, repression is lifted and efficient retransfer can occur throughout the population, a process resembling zygotic induction observed to occur with lysogenic bacteriophage [129].
TnsABC+E Pathway Targets Replication Forks.
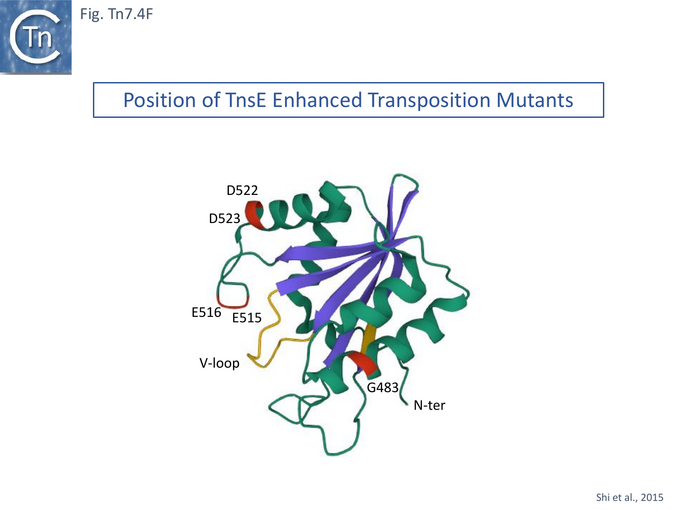
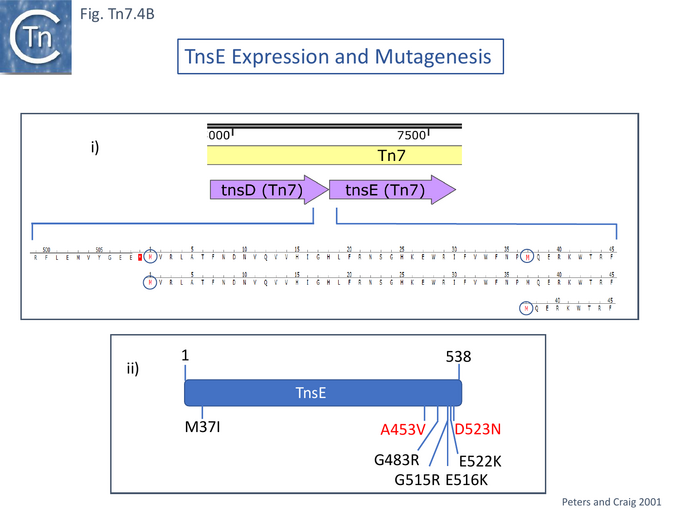
The question of the association of TnsABC+E transposition with the replication process has been investigated in some detail [122][130]. TnsE activity was assessed using two derivatives (Fig. Tn7.4Bi). Transposition was measured using the λ hop Assay with a miniTn7 and the transposition proteins were supplied in trans from a resident plasmid [122]. The native TnsE, as previously proposed, was found to be expressed from a GTG codon abutting the TnsD termination codon. A second derivative using the ATG codon at position 37 in TnsE was also cloned into an expression vector and was found to support lower levels of transposition than the full length protein. High-activity TnsE mutants (Fig. Tn7.4Bii) were identified after hydroxylamine mutagenesis using the papillation assay. TnsE (A453V) and TnsE (D523N) were isolated as a double mutant and each alone showed a 10X stimulation while the double mutant was almost 10X higher than each single mutant. Additional double mutants were also obtained using TnsE (A453V) as a seed.
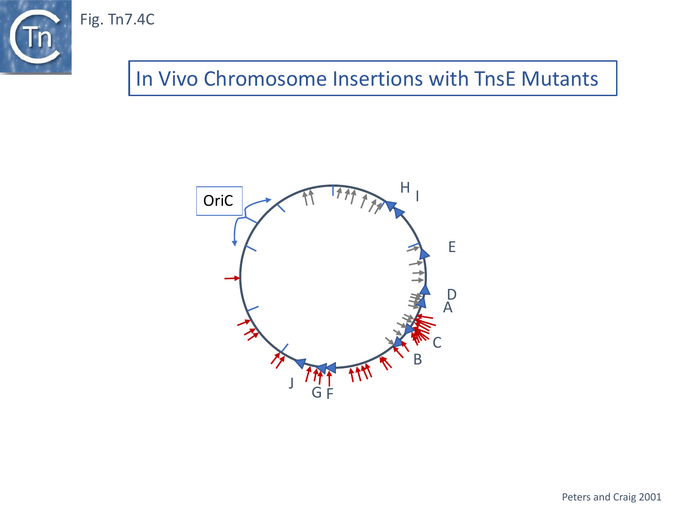
The mutants were tested for the partition of insertions between the chromosome and a resident pOXGen plasmid: wildtype TnsE directed the majority of insertions into the plasmid; each single mutant TnsE (A453V) or TnsE(D523N) yielded roughly equal numbers of insertions between plasmid and chromosome; while the double mutants TnsE (A453V/G515R) and TnsE (A453V/D523N) generated chromosomal insertions almost exclusively. Remarkably, those insertions into the pOXGen plasmid occurred in a single orientation and the chromosome insertions promoted by TnsE mutants occurred in opposite orientations on either side of OriC (Fig. Tn7.4C) except in the region of the terminus where they appear to occur in either orientation where replication forks arrive from both directions.
As an explanation for this behavior, it was proposed that, at low wildtype TnsE levels, the normal target structures in the cell are limiting. This is suggested by the fact that TnsE-mediated transposition is promoted by stimulation of conjugation [124] or double strand DNA breaks [106] and, in unpublished experiments (J.E. Peters and N.L. Craig, unpubl.), that this increase did not occur at high TnsE levels. Moreover, a quantitative difference in transposition “frequency” was observed between the λ hop Assay and the mating-out assay. Increasing TnsE expression of use the TnsE mutants can stimulate transposition by about 100 fold in the λ hop Assay but only marginally (8-10 fold) in the mating-out assay. This was interpreted as indicating that the TnsE mutant uses the same conjugal target(s) as the wildtype protein but find (additional) chromosome targets to achieve the very high transposition frequencies observed in the λ hop Assay [122].
TnsE Interacts Directly with 3’ Recessed Duplex DNA
Gel mobility shift assays revealed that TnsE bound double strand (DS) rather than single strand DNA and that the TnsE double mutants showed stronger binding. Moreover, the mutant TnsE generated a number of retarded bands on 5’ (Fig. Tn7.4Di) and on 3’ (Fig. Tn7.4Diii) recessed DS DNA but showed significant preference for the 3’ recessed end substrate (compared to the 5’ recessed substrate). More complex patterns and a stronger affinity were observed on branched DNA substrates resembling replication forks with either a 5’ (Fig. Tn7.4Dii) or 3’ (Fig. Tn7.4Div) single strand branches. The significantly higher TnsE affinity for 3’ recessed ends was postulated to reflect a preference for the lagging (discontinuous) strand at the replication fork and the idea arose that insertion orientation was directed by lagging strand synthesis [122][131] (Fig. Tn7.4Dv and vi). The reason why this was revealed only with high-activity TnsE mutants or increased levels of the wildtype protein is presumably because the excess “activity” allows less favourable target structures to be used.
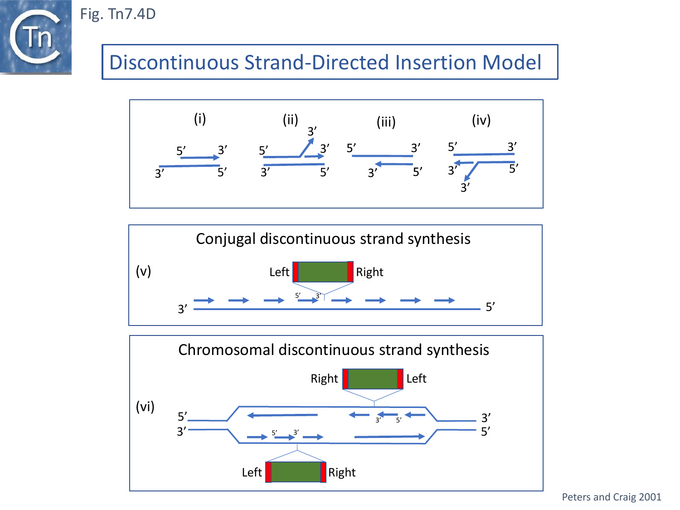
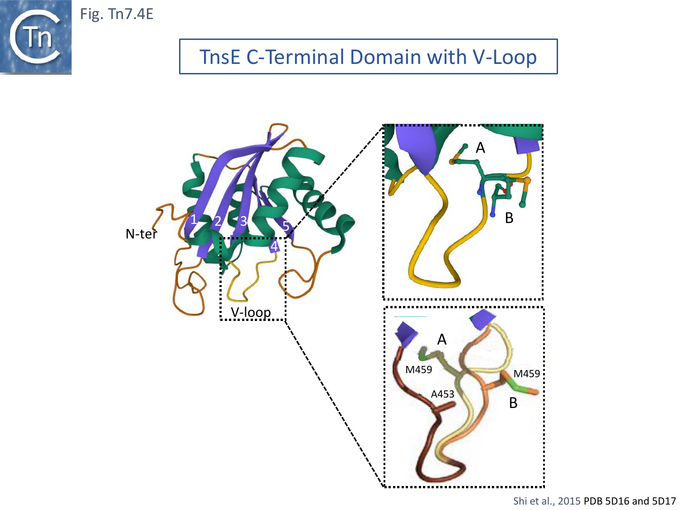
Further detailed understanding of how TnsE may interact with DNA in the TnsABC+E pathway was obtained from structural studies [132]. Partial trypsin proteolysis generated two major fragments of 48 and 25kDa: the smaller represented the TnsE C-terminus (CTD). Since it had been observed that TnsE mutants which enhance transposition clustered in the C-terminus of the protein [122] (Fig. Tn.7.4Bii), the CTD crystal structure (Fig. Tn7.4E) revealed a novel fold with a central five-stranded antiparallel-sheet surrounded by five helices (Fig. Tn7.4E left) which appears to be conserved in TnsE examples from a number of bacterial species [132]. β 3 and 4 are joined by a V-loop with arms composed of V451-D455 and V457-S461. In the wildtype protein this is “toggled” between two configurations (Fig. Tn7.4E, A and B, right).
However, in the mutants (TnsE-CTD(A453V+D523N)), the V-loop appears to be locked into a single configuration due to the A453V mutation. It was proposed that the gain of function mutants enhance transposition by two complementary mechanisms: the TnsEA453V mutation is located in the buried arm of the V-loop (V451-D455) and locks the other arm (V457-S461) into a configuration which favors substrate interactions while the TnsED523N mutation, located on the surface, extends a positively charged area increasing the DNA binding surface. Similar arguments explain the behavior of the other gain-of-activity mutants G483R, G515R, E516K and E522K (Fig. Tn7.4F).
Moreover, competition experiments in which the different TnsE mutants were premixed with labelled 3’ recessed DNA fragments and then challenged with unlabelled DNA revealed that, while the single mutants, TnsE (A453V) and TnsE (D523N) poorly resisted the challenge, DNA complexes with the double mutants were significantly more resistant to challenge.
TnsE also Interacts with the Replication Processivity Factor DnaN
Since the TnsD targeting protein relies in part on additional host factors in activating transposition [59], it was suggested that TnsE-mediated transposition might also involve host factors, in this case, proteins associated with discontinuous DNA replication. It was suggested that the target recognized by TnsE in the replication process could be the DNA replication processivity factor or DNA β clamp, DnaN [130] which is enriched on discontinuously replicating DNA (see [133]) and interacts with a number of proteins associated with DNA repair such as E. coli DNA polymerase IV [134] and MutL/S [135] and Okazaki fragment processing (see [136]).
Proteins which interact with sliding clamp proteins often exhibit a specific motif: QL(S/D)LF [137] or QLxLxL [138]. An alignment of a collection of TnsE-related proteins see [139][140] and Diversity below) revealed a moderately good consensus sequence which resembles the β clamp binding motif (Fig. Tn7.4Gi).
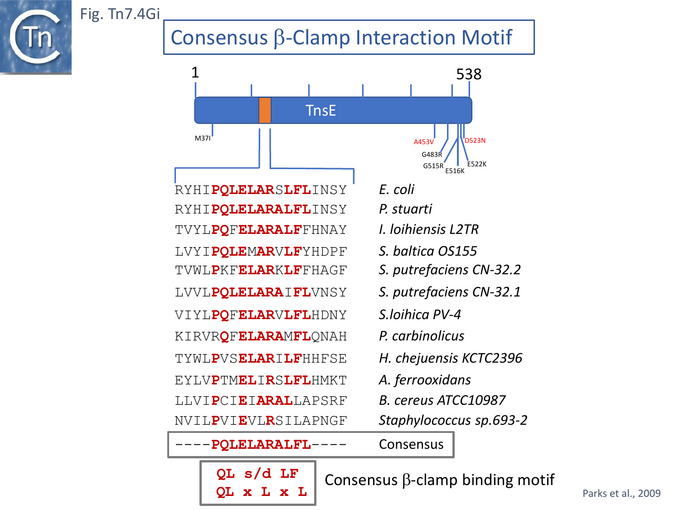
This was tested genetically using the yeast two-hybrid system with TnsE and β-clamp derivatives fused to the yeast GAL4 transcription activation and DNA binding domains, respectively [141] and, as readout, the β-galactosidase (β-gal) activity in a reporter strain containing the lacZ gene under control of a GAL4 promoter. With TnsE as “bait” and the β subunit as “prey” a significant signal was obtained indicating that TnsE and β-clamp do indeed interact [130]. The interaction was confirmed using a protein gel mobility shift with purified P32- labelled β-clamp and purified TnsE and by surface plasmon resonance (SPR) capable of monitoring relatively weak protein-protein interactions. To determine whether the potential β-clamp interaction motif is involved, a series of individual mutations changing six of the conserved consensus residues to alanine (Fig. Tn7.4Gii) exhibited a reduced signal in the yeast two hybrid system and also when analysed by SPR. They also resulted in a significantly reduced transposition frequency (4-40 fold). Moreover, increased expression of β generated significantly higher levels of TnsABC+E transposition but had no effect on TnsABC+D transposition.
TnsABC+E in vitro Transposition and the Effect of the β clamp
To further investigate the association with the β clamp, an in vitro TnsABC+E transposition system was developed using target plasmid of just over 6kb carrying a gap of 20 bp single-stranded DNA and a Tn7 donor plasmid with a conditional replication system requiring the ❁ protein (see [142]). Using purified TnsABC+E components, only the hyperactive TnsE mutants [122] generated detectable products after transformation into an appropriate host strain (without the ❁ protein) in a reaction which required the gapped plasmid target. There was no effect on transposition frequency between the gapped and a non-gapped target in the TnABC* core untargeted reaction which uses a TnsD/E independent TnsC mutant, TnsC(A225V) (Fig. Tn7.3E) [99] and, as expected [122], insertion appeared random occurring in both orientations in both the gapped and ungapped target. A number of these insertions were sequenced and shown to carry the expected 5bp flanking direct target repeats characteristic of Tn7 transposition.
Use of the TnsE mutants (TnsE*) increased transposition frequency between 300 and 1000-fold and, significantly, also appeared to occur randomly in the gapped target (Fig. Tn7.4Hi) in contrast to the insertion pattern observed in vivo [122].
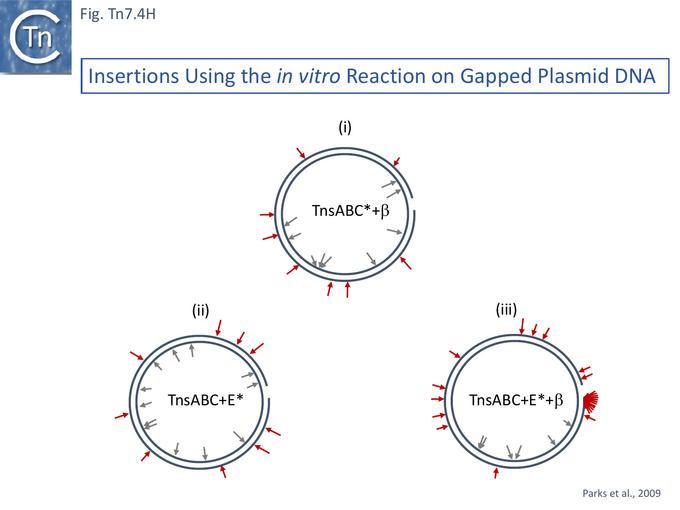
The effect of the β clamp was investigated by addition of a purified minimal clamp loader preparation (γ3δδ’) and the β clamp which loads specifically onto the gap [143] in a single orientation on the strand with the free 3’ end [133][144]. This had no effect on the distribution of insertions obtained with the TnABC* reaction which remained random (Fig. Tn7.4Hii) but had a significant effect on the TnsABC+E* reaction products (Fig. Tn7.4Hiii): 80% of the insertion events occurring in a single orientation, and 40% of them were were located at a single base pair junction proximal to the single-stranded DNA gap and 66 bp from the 3’ end [130].
Indeed, very high levels of TnsE* were found to induce SOS and the induction was reduced or eliminated in mutants within the proposed β clamp interaction domain (Fig. Tn7.4Gii).
The results establish a tight link between TnsABC+E transposition and DNA replication via DnaN as depicted in the model shown in Fig. Tn7.4I.
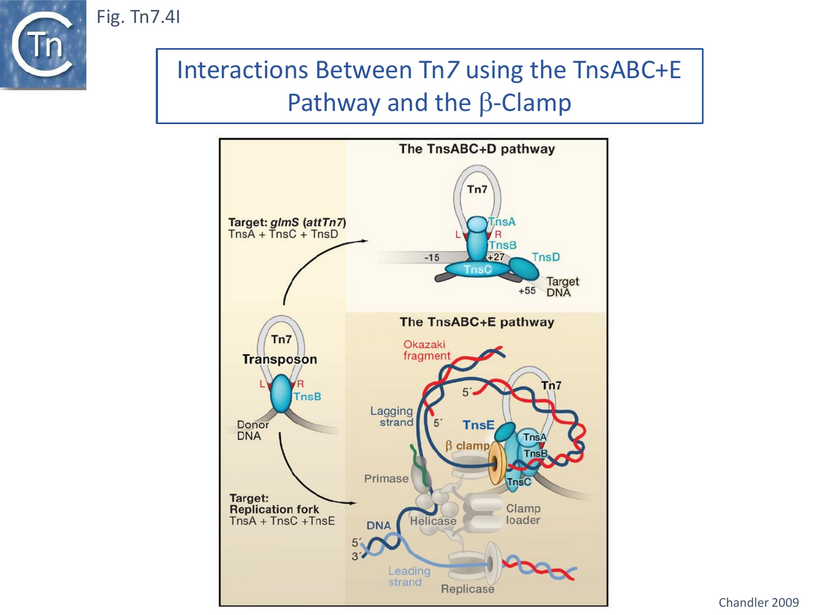
Immunity
in process, please check back later
in process, please check back later
Tn7 Integron
in process, please check back later
Tn7 Family Diversity
in process, please check back later
Bibliography
- ↑ Datta N, Barth PT . Compatibility properties of R483, a member of the I plasmid complex. - J Bacteriol: 1976 Mar, 125(3);796-9 [PubMed:130377] [DOI]
- ↑ 2.0 2.1 2.2 Barth PT, Datta N, Hedges RW, Grinter NJ . Transposition of a deoxyribonucleic acid sequence encoding trimethoprim and streptomycin resistances from R483 to other replicons. - J Bacteriol: 1976 Mar, 125(3);800-10 [PubMed:767328] [DOI]
- ↑
Chandler M, Silver L, Caro L . Suppression of an Escherichia coli dnaA mutation by the integrated R factor R100.1: origin of chromosome replication during exponential growth. - J Bacteriol: 1977 Aug, 131(2);421-30 [PubMed:328481]
[DOI]
- ↑ Bird RE, Chandler M, Caro L . Suppression of an Escherichia coli dnaA mutation by the integrated R factor R.100.1: Change of chromosome replication origin in synchronized cultures. - J Bacteriol: 1976 Jun, 126(3);1215-23 [PubMed:780344] [DOI]
- ↑ Chandler M, Silver L, Roth Y, Caro L . Chromosome replication in an Hfr strain of Escherichia coli. - J Mol Biol: 1976 Jun 25, 104(2);517-23 [PubMed:781291] [DOI]
- ↑ Nishimura Y, Caro L, Berg CM, Hirota Y . Chromosome replication in Escherichia coli. IV. Control of chromosome replication and cell division by an integrated episome. - J Mol Biol: 1971 Feb 14, 55(3);441-56 [PubMed:4927942] [DOI]
- ↑ Lederberg EM. Plasmid reference center registry of transposon (Tn) allocations through July 1981. Gene. 1981;16:59–61.
- ↑ Campbell A, Berg DE, Botstein D, Lederberg EM, Novick RP, Starlinger P, Szybalski W. Nomenclature of transposable elements in prokaryotes. Gene. 1979;5:197–206.
- ↑ 9.0 9.1 9.2 Lichtenstein C, Brenner S . Site-specific properties of Tn7 transposition into the E. coli chromosome. - Mol Gen Genet: 1981, 183(2);380-7 [PubMed:6276688] [DOI]
- ↑ 10.0 10.1 10.2 Lichtenstein C, Brenner S . Unique insertion site of Tn7 in the E. coli chromosome. - Nature: 1982 Jun 17, 297(5867);601-3 [PubMed:6283361] [DOI]
- ↑ Gosti-Testu F, Brevet J . [Determination of the terminal sequences of the transposon Tn7]. - C R Seances Acad Sci III: 1982 Jan 25, 294(4);193-6 [PubMed:6282401]
- ↑ 12.0 12.1 12.2 12.3 12.4 12.5 12.6 12.7 Arciszewska LK, Drake D, Craig NL . Transposon Tn7. cis-Acting sequences in transposition and transposition immunity. - J Mol Biol: 1989 May 5, 207(1);35-52 [PubMed:2544738] [DOI]
- ↑ 13.0 13.1 13.2 Gay NJ, Tybulewicz VL, Walker JE . Insertion of transposon Tn7 into the Escherichia coli glmS transcriptional terminator. - Biochem J: 1986 Feb 15, 234(1);111-7 [PubMed:3010949] [DOI]
- ↑ DeBoy RT, Craig NL . Target site selection by Tn7: attTn7 transcription and target activity. - J Bacteriol: 2000 Jun, 182(11);3310-3 [PubMed:10809719] [DOI]
- ↑ 15.0 15.1 15.2 Barth PT, Grinter NJ . Map of plasmid RP4 derived by insertion of transposon C. - J Mol Biol: 1977 Jul 5, 113(3);455-74 [PubMed:328901] [DOI]
- ↑ 16.0 16.1 Barth PT, Grinter NJ, Bradley DE . Conjugal transfer system of plasmid RP4: analysis by transposon 7 insertion. - J Bacteriol: 1978 Jan, 133(1);43-52 [PubMed:338595] [DOI]
- ↑ 17.0 17.1 Moore RJ, Krishnapillai V . Tn7 and Tn501 Insertions into Pseudomonas aeruginosa plasmid R91-5: mapping of two transfer regions. - J Bacteriol: 1982 Jan, 149(1);276-83 [PubMed:6119306] [DOI]
- ↑ Hernalsteens JP, De Greve H, Van Montagu M, Schell J . Mutagenesis by insertion of the drug resistance transposon Tn7 applied to the Ti plasmid of Agrobacterium tumefaciens. - Plasmid: 1978 Feb, 1(2);218-25 [PubMed:748948] [DOI]
- ↑ Thomson JA, Hendson M, Magnes RM . Mutagenesis by insertion of drug resistance transposon Tn7 into a vibrio species. - J Bacteriol: 1981 Oct, 148(1);374-8 [PubMed:6270064] [DOI]
- ↑ Thomasian SK, Voll MJ . Specificity of transposition of Tn7 in Vibrio parahaemolyticus. - Plasmid: 1989 Jul, 22(1);82-5 [PubMed:2550986] [DOI]
- ↑ Caruso M, Shapiro JA . Interactions of Tn7 and temperate phage F116L of Pseudomonas aeruginosa. - Mol Gen Genet: 1982, 188(2);292-8 [PubMed:6296632] [DOI]
- ↑ Ely B . Transposition of Tn7 occurs at a single site on the Caulobacter crescentus chromosome. - J Bacteriol: 1982 Aug, 151(2);1056-8 [PubMed:6284700] [DOI]
- ↑ Oppon JC, Sarnovsky RJ, Craig NL, Rawlings DE . A Tn7-like transposon is present in the glmUS region of the obligately chemoautolithotrophic bacterium Thiobacillus ferrooxidans. - J Bacteriol: 1998 Jun, 180(11);3007-12 [PubMed:9603897] [DOI]
- ↑ 24.0 24.1 Nnalue NA . Tn7 inserts in both orientations at a single chromosomal location and apparently forms cointegrates in Pasteurella multocida. - Mol Microbiol: 1990 Jan, 4(1);107-17 [PubMed:2157128] [DOI]
- ↑ 25.0 25.1 Hauer B, Shapiro JA. Control of Tn7 transposition. MolGenGenet. 1984;194:149–158.
- ↑ 26.0 26.1 26.2 Smith GM, Jones P . Effects of deletions in transposon Tn7 on its frequency of transposition. - J Bacteriol: 1984 Mar, 157(3);962-4 [PubMed:6321449] [DOI]
- ↑ 27.0 27.1 Gosti-Testu F, Norris V, Brevet J . Restriction map of Tn7. - Plasmid: 1983 Jul, 10(1);96-9 [PubMed:6312477] [DOI]
- ↑ 28.0 28.1 Ouartsi A, Borowski D, Brevet J . Genetic analysis of Tn7 transposition. - Mol Gen Genet: 1985, 198(2);221-7 [PubMed:2984518] [DOI]
- ↑ 29.0 29.1 29.2 Smith GM, Jones P . Tn7 transposition: a multigene process. Identification of a regulatory gene product. - Nucleic Acids Res: 1986 Oct 24, 14(20);7915-27 [PubMed:3022239] [DOI]
- ↑ 30.0 30.1 30.2 30.3 30.4 Brevet J, Faure F, Borowski D . Tn7-encoded proteins. - Mol Gen Genet: 1985, 201(2);258-64 [PubMed:3003528] [DOI]
- ↑ 31.0 31.1 31.2 31.3 31.4 31.5 31.6 31.7 31.8 31.9 Rogers M, Ekaterinaki N, Nimmo E, Sherratt D . Analysis of Tn7 transposition. - Mol Gen Genet: 1986 Dec, 205(3);550-6 [PubMed:3031432] [DOI]
- ↑ Flores CC, Cotterill S, Lichtenstein CP . Overproduction of four functionally active proteins, TnsA, B, C, and D, required for Tn7 transposition to its attachment site, attTn7. - Plasmid: 1992 Jul, 28(1);80-5 [PubMed:1325658] [DOI]
- ↑ 33.0 33.1 33.2 Hauer B, Shapiro JA. Control of Tn7 transposition. Molec Gen Genet. 1984 Apr;194(1-2):149–158.
- ↑ 34.0 34.1 Galas DJ, Chandler M. Structure and stability of Tn9-mediated cointegrates. J Mol Biol. 1982 Jan;154(2):245–272.
- ↑ 35.0 35.1 Guyer MS, Reed RR, Steitz JA, Low KB . Identification of a sex-factor-affinity site in E. coli as gamma delta. - Cold Spring Harb Symp Quant Biol: 1981, 45 Pt 1;135-40 [PubMed:6271456] [DOI]
- ↑ Yin JC, Krebs MP, Reznikoff WS . Effect of dam methylation on Tn5 transposition. - J Mol Biol: 1988 Jan 5, 199(1);35-45 [PubMed:2451025] [DOI]
- ↑ Foster TJ, Davis MA, Roberts DE, Takeshita K, Kleckner N . Genetic organization of transposon Tn10. - Cell: 1981 Jan, 23(1);201-13 [PubMed:6260375] [DOI]
- ↑ 38.0 38.1 Huisman O, Kleckner N . A new generalizable test for detection of mutations affecting Tn10 transposition. - Genetics: 1987 Jun, 116(2);185-9 [PubMed:3038669] [DOI]
- ↑ 39.0 39.1 Krebs MP, Reznikoff WS. Use of a Tn5 derivative that creates lacZ translational fusions to obtain a transposition mutant. Gene. 1988;63:277–285.
- ↑ 40.0 40.1 40.2 40.3 40.4 40.5 Waddell CS, Craig NL . Tn7 transposition: two transposition pathways directed by five Tn7-encoded genes. - Genes Dev: 1988 Feb, 2(2);137-49 [PubMed:2834269] [DOI]
- ↑ Fellay R, Krisch HM, Prentki P, Frey J. Omegon-Km: a transposable element designed for in vivo insertional mutagenesis and cloning of genes in gram-negative bacteria. Gene. 1989;76:215–226.
- ↑ 42.0 42.1 42.2 42.3 42.4 42.5 Orle KA, Craig NL. Identification of transposition proteins encoded by the bacterial transposon Tn7 [corrected and republished article originally printed in Gene 1990 Nov 30;96(1):1-7]. Gene. 1991;104:125–131.
- ↑ 43.0 43.1 43.2 43.3 Flores C, Qadri MI, Lichtenstein C . DNA sequence analysis of five genes; tnsA, B, C, D and E, required for Tn7 transposition. - Nucleic Acids Res: 1990 Feb 25, 18(4);901-11 [PubMed:2156235] [DOI]
- ↑ Huber M, Faure G, Laass S, Kolbe E, Seitz K, Wehrheim C, Wolf YI, Koonin EV, Soppa J . Translational coupling via termination-reinitiation in archaea and bacteria. - Nat Commun: 2019 Sep 5, 10(1);4006 [PubMed:31488843] [DOI]
- ↑ Rex G, Surin B, Besse G, Schneppe B, McCarthy JE . The mechanism of translational coupling in Escherichia coli. Higher order structure in the atpHA mRNA acts as a conformational switch regulating the access of de novo initiating ribosomes. - J Biol Chem: 1994 Jul 8, 269(27);18118-27 [PubMed:7517937]
- ↑ 46.0 46.1 46.2 McKown RL, Orle KA, Chen T, Craig NL . Sequence requirements of Escherichia coli attTn7, a specific site of transposon Tn7 insertion. - J Bacteriol: 1988 Jan, 170(1);352-8 [PubMed:2826397] [DOI]
- ↑ 47.0 47.1 Qadri MI, Flores CC, Davis AJ, Lichtenstein CP . Genetic analysis of attTn7, the transposon Tn7 attachment site in Escherichia coli, using a novel M13-based transduction assay. - J Mol Biol: 1989 May 5, 207(1);85-98 [PubMed:2544739] [DOI]
- ↑ Gringauz E, Orle KA, Waddell CS, Craig NL. Recognition of Escherichia coli attTn7 by transposon Tn7: lack of specific sequence requirements at the point of Tn7 insertion [published erratum appears in J Bacteriol 1988 Aug;170(8):3792]. J Bacteriol. 1988;170:2832–2840.
- ↑ 49.0 49.1 Waddell CS, Craig NL . Tn7 transposition: recognition of the attTn7 target sequence. - Proc Natl Acad Sci U S A: 1989 Jun, 86(11);3958-62 [PubMed:2542960] [DOI]
- ↑ 50.0 50.1 Kubo KM, Craig NL . Bacterial transposon Tn7 utilizes two different classes of target sites. - J Bacteriol: 1990 May, 172(5);2774-8 [PubMed:2158980] [DOI]
- ↑ 51.0 51.1 51.2 51.3 51.4 51.5 51.6 Bainton R, Gamas P, Craig NL . Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. - Cell: 1991 May 31, 65(5);805-16 [PubMed:1645619] [DOI]
- ↑ Hagemann AT, Craig NL . Tn7 transposition creates a hotspot for homologous recombination at the transposon donor site. - Genetics: 1993 Jan, 133(1);9-16 [PubMed:8380272] [DOI]
- ↑ 53.0 53.1 53.2 53.3 53.4 53.5 Arciszewska LK, McKown RL, Craig NL . Purification of TnsB, a transposition protein that binds to the ends of Tn7. - J Biol Chem: 1991 Nov 15, 266(32);21736-44 [PubMed:1657979]
- ↑ 54.0 54.1 54.2 54.3 54.4 54.5 54.6 54.7 Gamas P, Craig NL . Purification and characterization of TnsC, a Tn7 transposition protein that binds ATP and DNA. - Nucleic Acids Res: 1992 May 25, 20(10);2525-32 [PubMed:1317955] [DOI]
- ↑ 55.00 55.01 55.02 55.03 55.04 55.05 55.06 55.07 55.08 55.09 55.10 Bainton RJ, Kubo KM, Feng JN, Craig NL . Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. - Cell: 1993 Mar 26, 72(6);931-43 [PubMed:8384534] [DOI]
- ↑ 56.0 56.1 56.2 56.3 Kuduvalli PN, Rao JE, Craig NL . Target DNA structure plays a critical role in Tn7 transposition. - EMBO J: 2001 Feb 15, 20(4);924-32 [PubMed:11179236] [DOI]
- ↑ 57.0 57.1 57.2 57.3 Rao JE, Miller PS, Craig NL . Recognition of triple-helical DNA structures by transposon Tn7. - Proc Natl Acad Sci U S A: 2000 Apr 11, 97(8);3936-41 [PubMed:10737770] [DOI]
- ↑ 58.0 58.1 Rao JE, Craig NL . Selective recognition of pyrimidine motif triplexes by a protein encoded by the bacterial transposon Tn7. - J Mol Biol: 2001 Apr 13, 307(5);1161-70 [PubMed:11292332] [DOI]
- ↑ 59.0 59.1 59.2 Sharpe PL, Craig NL . Host proteins can stimulate Tn7 transposition: a novel role for the ribosomal protein L29 and the acyl carrier protein. - EMBO J: 1998 Oct 1, 17(19);5822-31 [PubMed:9755182] [DOI]
- ↑ Maekawa T, Yanagihara K, Ohtsubo E . Specific nicking at the 3' ends of the terminal inverted repeat sequences in transposon Tn3 by transposase and an E. coli protein ACP. - Genes Cells: 1996 Nov, 1(11);1017-30 [PubMed:9077464] [DOI]
- ↑ Rock CO, Cronan JE Jr . Acyl-acyl carrier protein synthetase from Escherichia coli. - Methods Enzymol: 1981, 71 Pt C;163-8 [PubMed:7024728] [DOI]
- ↑ Magnuson K, Jackowski S, Rock CO, Cronan JE Jr . Regulation of fatty acid biosynthesis in Escherichia coli. - Microbiol Rev: 1993 Sep, 57(3);522-42 [PubMed:8246839] [DOI]
- ↑ 63.0 63.1 63.2 63.3 Gary PA, Biery MC, Bainton RJ, Craig NL . Multiple DNA processing reactions underlie Tn7 transposition. - J Mol Biol: 1996 Mar 29, 257(2);301-16 [PubMed:8609625] [DOI]
- ↑ 64.0 64.1 Polard P, Chandler M . An in vivo transposase-catalyzed single-stranded DNA circularization reaction. - Genes Dev: 1995 Nov 15, 9(22);2846-58 [PubMed:7590258] [DOI]
- ↑ 65.0 65.1 Tang Y, Cotterill S, Lichtenstein CP. Genetic analysis of the terminal 8-bp inverted repeats of transposon Tn7. Gene. 1995 Aug;162(1):41–46.
- ↑ Derbyshire KM, Hwang L, Grindley ND . Genetic analysis of the interaction of the insertion sequence IS903 transposase with its terminal inverted repeats. - Proc Natl Acad Sci U S A: 1987 Nov, 84(22);8049-53 [PubMed:2825175] [DOI]
- ↑ Lewis LA, Cylin E, Lee HK, Saby R, Wong W, Grindley ND . The left end of IS2: a compromise between transpositional activity and an essential promoter function that regulates the transposition pathway. - J Bacteriol: 2004 Feb, 186(3);858-65 [PubMed:14729714] [DOI]
- ↑ Lewis LA, Gadura N, Greene M, Saby R, Grindley ND . The basis of asymmetry in IS2 transposition. - Mol Microbiol: 2001 Nov, 42(4);887-901 [PubMed:11737634] [DOI]
- ↑ 69.0 69.1 69.2 69.3 May EW, Craig NL . Switching from cut-and-paste to replicative Tn7 transposition. - Science: 1996 Apr 19, 272(5260);401-4 [PubMed:8602527] [DOI]
- ↑ 70.0 70.1 Shapiro JA . Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. - Proc Natl Acad Sci U S A: 1979 Apr, 76(4);1933-7 [PubMed:287033] [DOI]
- ↑ Hsu M, Berg P . Altering the specificity of restriction endonuclease: effect of replacing Mg2+ with Mn2+. - Biochemistry: 1978 Jan 10, 17(1);131-38 [PubMed:201281] [DOI]
- ↑ Vermote CL, Halford SE . EcoRV restriction endonuclease: communication between catalytic metal ions and DNA recognition. - Biochemistry: 1992 Jul 7, 31(26);6082-9 [PubMed:1627551] [DOI]
- ↑ Bennett SP, Halford SE . Recognition of DNA by type II restriction enzymes. - Curr Top Cell Regul: 1989, 30;57-104 [PubMed:2695290] [DOI]
- ↑ Baker TA, Luo L . Identification of residues in the Mu transposase essential for catalysis. - Proc Natl Acad Sci U S A: 1994 Jul 5, 91(14);6654-8 [PubMed:7912831] [DOI]
- ↑ 75.0 75.1 75.2 Sarnovsky RJ, May EW, Craig NL . The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. - EMBO J: 1996 Nov 15, 15(22);6348-61 [PubMed:8947057]
- ↑ 76.0 76.1 Biery MC, Lopata M, Craig NL . A minimal system for Tn7 transposition: the transposon-encoded proteins TnsA and TnsB can execute DNA breakage and joining reactions that generate circularized Tn7 species. - J Mol Biol: 2000 Mar 17, 297(1);25-37 [PubMed:10704304] [DOI]
- ↑ 77.0 77.1 77.2 77.3 Tang Y, Lichtenstein C, Cotterill S . Purification and characterisation of the TnsB protein of Tn7: a transposition protein that binds to the ends of Tn7. - Nucleic Acids Res: 1991 Jun 25, 19(12);3395-402 [PubMed:1648205] [DOI]
- ↑ 78.0 78.1 78.2 McKown RL, Waddell CS, Arciszewska LK, Craig NL . Identification of a transposon Tn7-dependent DNA-binding activity that recognizes the ends of Tn7. - Proc Natl Acad Sci U S A: 1987 Nov, 84(22);7807-11 [PubMed:2825163] [DOI]
- ↑ 79.0 79.1 79.2 79.3 79.4 79.5 79.6 Arciszewska LK, Craig NL . Interaction of the Tn7-encoded transposition protein TnsB with the ends of the transposon. - Nucleic Acids Res: 1991 Sep 25, 19(18);5021-9 [PubMed:1656385] [DOI]
- ↑ Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. ProcNatlAcadSciUSA. 1993;90(14):6498–6502.
- ↑ Beese LS, Steitz TA . Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. - EMBO J: 1991 Jan, 10(1);25-33 [PubMed:1989886] [DOI]
- ↑ Dahm SC, Uhlenbeck OC . Role of divalent metal ions in the hammerhead RNA cleavage reaction. - Biochemistry: 1991 Oct 1, 30(39);9464-9 [PubMed:1716459] [DOI]
- ↑ Piccirilli JA, Vyle JS, Caruthers MH, Cech TR . Metal ion catalysis in the Tetrahymena ribozyme reaction. - Nature: 1993 Jan 7, 361(6407);85-8 [PubMed:8421499] [DOI]
- ↑ Engelman A, Craigie R . Efficient magnesium-dependent human immunodeficiency virus type 1 integrase activity. - J Virol: 1995 Sep, 69(9);5908-11 [PubMed:7637039] [DOI]
- ↑ 85.0 85.1 Kaczmarska Z, Czarnocki-Cieciura M, Górecka-Minakowska KM, Wingo RJ, Jackiewicz J, Zajko W, Poznański JT, Rawski M, Grant T, Peters JE, Nowotny M . Structural basis of transposon end recognition explains central features of Tn7 transposition systems. - Mol Cell: 2022 Jul 21, 82(14);2618-2632.e7 [PubMed:35654042] [DOI]
- ↑ Montaño SP, Pigli YZ, Rice PA . The μ transpososome structure sheds light on DDE recombinase evolution. - Nature: 2012 Nov 15, 491(7424);413-7 [PubMed:23135398] [DOI]
- ↑ 87.0 87.1 87.2 87.3 87.4 87.5 87.6 87.7 Hickman AB, Li Y, Mathew SV, May EW, Craig NL, Dyda F . Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. - Mol Cell: 2000 Jun, 5(6);1025-34 [PubMed:10911996] [DOI]
- ↑ Wah DA, Hirsch JA, Dorner LF, Schildkraut I, Aggarwal AK . Structure of the multimodular endonuclease FokI bound to DNA. - Nature: 1997 Jul 3, 388(6637);97-100 [PubMed:9214510] [DOI]
- ↑ Wah DA, Bitinaite J, Schildkraut I, Aggarwal AK . Structure of FokI has implications for DNA cleavage. - Proc Natl Acad Sci U S A: 1998 Sep 1, 95(18);10564-9 [PubMed:9724743] [DOI]
- ↑ Aggarwal AK, Wah DA . Novel site-specific DNA endonucleases. - Curr Opin Struct Biol: 1998 Feb, 8(1);19-25 [PubMed:9519292] [DOI]
- ↑ Pernstich C, Halford SE . Illuminating the reaction pathway of the FokI restriction endonuclease by fluorescence resonance energy transfer. - Nucleic Acids Res: 2012 Feb, 40(3);1203-13 [PubMed:21993298] [DOI]
- ↑ 92.0 92.1 92.2 Stellwagen AE, Craig NL . Mobile DNA elements: controlling transposition with ATP-dependent molecular switches. - Trends Biochem Sci: 1998 Dec, 23(12);486-90 [PubMed:9868372] [DOI]
- ↑ 93.0 93.1 93.2 93.3 Lu F, Craig NL . Isolation and characterization of Tn7 transposase gain-of-function mutants: a model for transposase activation. - EMBO J: 2000 Jul 3, 19(13);3446-57 [PubMed:10880457] [DOI]
- ↑ Sheshberadaran H, Payne LG . Protein antigen-monoclonal antibody contact sites investigated by limited proteolysis of monoclonal antibody-bound antigen: protein "footprinting". - Proc Natl Acad Sci U S A: 1988 Jan, 85(1);1-5 [PubMed:2448767] [DOI]
- ↑ Sancar A, Hearst JE . Molecular matchmakers. - Science: 1993 Mar 5, 259(5100);1415-20 [PubMed:8451638] [DOI]
- ↑ 96.0 96.1 Stellwagen AE, Craig NL . Gain-of-function mutations in TnsC, an ATP-dependent transposition protein that activates the bacterial transposon Tn7. - Genetics: 1997 Mar, 145(3);573-85 [PubMed:9055068] [DOI]
- ↑ Hopkins JD, Clements M, Syvanen M . New class of mutations in Escherichia coli (uup) that affect precise excision of insertion elements and bacteriophage Mu growth. - J Bacteriol: 1983 Jan, 153(1);384-9 [PubMed:6294054] [DOI]
- ↑ Rhee S, Han Zj, Liu K, Miles HT, Davies DR . Structure of a triple helical DNA with a triplex-duplex junction. - Biochemistry: 1999 Dec 21, 38(51);16810-5 [PubMed:10606513] [DOI]
- ↑ 99.0 99.1 99.2 99.3 99.4 99.5 Stellwagen AE, Craig NL . Analysis of gain-of-function mutants of an ATP-dependent regulator of Tn7 transposition. - J Mol Biol: 2001 Jan 19, 305(3);633-42 [PubMed:11152618] [DOI]
- ↑ 100.0 100.1 100.2 100.3 100.4 Ronning DR, Li Y, Perez ZN, Ross PD, Hickman AB, Craig NL, Dyda F . The carboxy-terminal portion of TnsC activates the Tn7 transposase through a specific interaction with TnsA. - EMBO J: 2004 Aug 4, 23(15);2972-81 [PubMed:15257292] [DOI]
- ↑ 101.0 101.1 101.2 101.3 Skelding Z, Queen-Baker J, Craig NL . Alternative interactions between the Tn7 transposase and the Tn7 target DNA binding protein regulate target immunity and transposition. - EMBO J: 2003 Nov 3, 22(21);5904-17 [PubMed:14592987] [DOI]
- ↑ Skelding Z, Sarnovsky R, Craig NL . Formation of a nucleoprotein complex containing Tn7 and its target DNA regulates transposition initiation. - EMBO J: 2002 Jul 1, 21(13);3494-504 [PubMed:12093750] [DOI]
- ↑ Choi KY, Spencer JM, Craig NL . The Tn7 transposition regulator TnsC interacts with the transposase subunit TnsB and target selector TnsD. - Proc Natl Acad Sci U S A: 2014 Jul 15, 111(28);E2858-65 [PubMed:24982178] [DOI]
- ↑ 104.00 104.01 104.02 104.03 104.04 104.05 104.06 104.07 104.08 104.09 104.10 104.11 Shen Y, Gomez-Blanco J, Petassi MT, Peters JE, Ortega J, Guarné A . Structural basis for DNA targeting by the Tn7 transposon. - Nat Struct Mol Biol: 2022 Feb, 29(2);143-151 [PubMed:35173349] [DOI]
- ↑ 105.0 105.1 Arias-Palomo E, Berger JM . An Atypical AAA+ ATPase Assembly Controls Efficient Transposition through DNA Remodeling and Transposase Recruitment. - Cell: 2015 Aug 13, 162(4);860-71 [PubMed:26276634] [DOI]
- ↑ 106.0 106.1 106.2 106.3 106.4 106.5 106.6 Peters JE, Craig NL . Tn7 transposes proximal to DNA double-strand breaks and into regions where chromosomal DNA replication terminates. - Mol Cell: 2000 Sep, 6(3);573-82 [PubMed:11030337] [DOI]
- ↑ Bierne H, Michel B . When replication forks stop. - Mol Microbiol: 1994 Jul, 13(1);17-23 [PubMed:7984091] [DOI]
- ↑ Neylon C, Kralicek AV, Hill TM, Dixon NE . Replication termination in Escherichia coli: structure and antihelicase activity of the Tus-Ter complex. - Microbiol Mol Biol Rev: 2005 Sep, 69(3);501-26 [PubMed:16148308] [DOI]
- ↑ Hill TM, Marians KJ . Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. - Proc Natl Acad Sci U S A: 1990 Apr, 87(7);2481-5 [PubMed:2181438] [DOI]
- ↑ Mulcair MD, Schaeffer PM, Oakley AJ, Cross HF, Neylon C, Hill TM, Dixon NE . A molecular mousetrap determines polarity of termination of DNA replication in E. coli. - Cell: 2006 Jun 30, 125(7);1309-19 [PubMed:16814717] [DOI]
- ↑ Kuempel PL, Pelletier AJ, Hill TM . Tus and the terminators: the arrest of replication in prokaryotes. - Cell: 1989 Nov 17, 59(4);581-3 [PubMed:2510933] [DOI]
- ↑ Hill TM, Tecklenburg ML, Pelletier AJ, Kuempel PL . tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. - Proc Natl Acad Sci U S A: 1989 Mar, 86(5);1593-7 [PubMed:2646639] [DOI]
- ↑ Stellwagen AE, Craig NL . Avoiding self: two Tn7-encoded proteins mediate target immunity in Tn7 transposition. - EMBO J: 1997 Nov 17, 16(22);6823-34 [PubMed:9362496] [DOI]
- ↑ Louarn JM, Louarn J, François V, Patte J . Analysis and possible role of hyperrecombination in the termination region of the Escherichia coli chromosome. - J Bacteriol: 1991 Aug, 173(16);5097-104 [PubMed:1650344] [DOI]
- ↑ Bierne H, Ehrlich SD, Michel B . The replication termination signal terB of the Escherichia coli chromosome is a deletion hot spot. - EMBO J: 1991 Sep, 10(9);2699-705 [PubMed:1868840] [DOI]
- ↑ Haniford DB, Chelouche AR, Kleckner N . A specific class of IS10 transposase mutants are blocked for target site interactions and promote formation of an excised transposon fragment. - Cell: 1989 Oct 20, 59(2);385-94 [PubMed:2553270] [DOI]
- ↑ Galitski T, Roth JR . Pathways for homologous recombination between chromosomal direct repeats in Salmonella typhimurium. - Genetics: 1997 Jul, 146(3);751-67 [PubMed:9215885] [DOI]
- ↑ Corre J, Cornet F, Patte J, Louarn JM . Unraveling a region-specific hyper-recombination phenomenon: genetic control and modalities of terminal recombination in Escherichia coli. - Genetics: 1997 Nov, 147(3);979-89 [PubMed:9383046] [DOI]
- ↑ Kuzminov A . Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. - Microbiol Mol Biol Rev: 1999 Dec, 63(4);751-813, table of contents [PubMed:10585965] [DOI]
- ↑ Yang L, Handa N, Liu B, Dillingham MS, Wigley DB, Kowalczykowski SC . Alteration of χ recognition by RecBCD reveals a regulated molecular latch and suggests a channel-bypass mechanism for biological control. - Proc Natl Acad Sci U S A: 2012 Jun 5, 109(23);8907-12 [PubMed:22603793] [DOI]
- ↑ 121.0 121.1 Shi Q, Parks AR, Potter BD, Safir IJ, Luo Y, Forster BM, Peters JE . DNA damage differentially activates regional chromosomal loci for Tn7 transposition in Escherichia coli. - Genetics: 2008 Jul, 179(3);1237-50 [PubMed:18562643] [DOI]
- ↑ 122.0 122.1 122.2 122.3 122.4 122.5 122.6 122.7 122.8 Peters JE, Craig NL . Tn7 recognizes transposition target structures associated with DNA replication using the DNA-binding protein TnsE. - Genes Dev: 2001 Mar 15, 15(6);737-47 [PubMed:11274058] [DOI]
- ↑ Shi Q, Huguet-Tapia JC, Peters JE . Tn917 targets the region where DNA replication terminates in Bacillus subtilis, highlighting a difference in chromosome processing in the firmicutes. - J Bacteriol: 2009 Dec, 191(24);7623-7 [PubMed:19820088] [DOI]
- ↑ 124.0 124.1 124.2 Wolkow CA, DeBoy RT, Craig NL . Conjugating plasmids are preferred targets for Tn7. - Genes Dev: 1996 Sep 1, 10(17);2145-57 [PubMed:8804309] [DOI]
- ↑ Everett R, Willetts N . Cloning, mutation, and location of the F origin of conjugal transfer. - EMBO J: 1982, 1(6);747-53 [PubMed:6329701] [DOI]
- ↑ Lanka E, Wilkins BM . DNA processing reactions in bacterial conjugation. - Annu Rev Biochem: 1995, 64;141-69 [PubMed:7574478] [DOI]
- ↑ Wilkins BM, Hollom SE . Conjugational synthesis of F lac+ and Col I DNA in the presence of rifampicin and in Escherichia coli K12 mutants defective in DNA synthesis. - Mol Gen Genet: 1974, 134(2);143-56 [PubMed:4617160] [DOI]
- ↑ Chandler M, de la Cruz F, Dyda F, Hickman AB, Moncalian G, Ton-Hoang B . Breaking and joining single-stranded DNA: the HUH endonuclease superfamily. - Nat Rev Microbiol: 2013 Aug, 11(8);525-38 [PubMed:23832240] [DOI]
- ↑ JACOB F, WOLLMAN EL . [Processes of conjugation and recombination in Escherichia coli. I. Induction by conjugation or zygotic induction]. - Ann Inst Pasteur (Paris): 1956 Oct, 91(4);486-510 [PubMed:13373067]
- ↑ 130.0 130.1 130.2 130.3 Parks AR, Li Z, Shi Q, Owens RM, Jin MM, Peters JE . Transposition into replicating DNA occurs through interaction with the processivity factor. - Cell: 2009 Aug 21, 138(4);685-95 [PubMed:19703395] [DOI]
- ↑ Peters JE, Craig NL . Tn7: smarter than we thought. - Nat Rev Mol Cell Biol: 2001 Nov, 2(11);806-14 [PubMed:11715047] [DOI]
- ↑ 132.0 132.1 Shi Q, Straus MR, Caron JJ, Wang H, Chung YS, Guarné A, Peters JE . Conformational toggling controls target site choice for the heteromeric transposase element Tn7. - Nucleic Acids Res: 2015 Dec 15, 43(22);10734-45 [PubMed:26384427] [DOI]
- ↑ 133.0 133.1 Johnson A, O'Donnell M . Cellular DNA replicases: components and dynamics at the replication fork. - Annu Rev Biochem: 2005, 74;283-315 [PubMed:15952889] [DOI]
- ↑ Bunting KA, Roe SM, Pearl LH . Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. - EMBO J: 2003 Nov 3, 22(21);5883-92 [PubMed:14592985] [DOI]
- ↑ López de Saro FJ, Marinus MG, Modrich P, O'Donnell M . The beta sliding clamp binds to multiple sites within MutL and MutS. - J Biol Chem: 2006 May 19, 281(20);14340-9 [PubMed:16546997] [DOI]
- ↑ López de Saro FJ . Regulation of interactions with sliding clamps during DNA replication and repair. - Curr Genomics: 2009 May, 10(3);206-15 [PubMed:19881914] [DOI]
- ↑ Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA . A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. - Proc Natl Acad Sci U S A: 2001 Sep 25, 98(20);11627-32 [PubMed:11573000] [DOI]
- ↑ Wijffels G, Dalrymple BP, Prosselkov P, Kongsuwan K, Epa VC, Lilley PE, Jergic S, Buchardt J, Brown SE, Alewood PF, Jennings PA, Dixon NE . Inhibition of protein interactions with the beta 2 sliding clamp of Escherichia coli DNA polymerase III by peptides from beta 2-binding proteins. - Biochemistry: 2004 May 18, 43(19);5661-71 [PubMed:15134440] [DOI]
- ↑ Parks AR, Peters JE . Tn7 elements: engendering diversity from chromosomes to episomes. - Plasmid: 2009 Jan, 61(1);1-14 [PubMed:18951916] [DOI]
- ↑ Parks AR, Peters JE . Transposon Tn7 is widespread in diverse bacteria and forms genomic islands. - J Bacteriol: 2007 Mar, 189(5);2170-3 [PubMed:17194796] [DOI]
- ↑ Fields S, Song O . A novel genetic system to detect protein-protein interactions. - Nature: 1989 Jul 20, 340(6230);245-6 [PubMed:2547163] [DOI]
- ↑ Herrero M, de Lorenzo V, Timmis KN . Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. - J Bacteriol: 1990 Nov, 172(11);6557-67 [PubMed:2172216] [DOI]
- ↑ Leu FP, Hingorani MM, Turner J, O'Donnell M . The delta subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. - J Biol Chem: 2000 Nov 3, 275(44);34609-18 [PubMed:10924523] [DOI]
- ↑ Georgescu RE, Kim SS, Yurieva O, Kuriyan J, Kong XP, O'Donnell M . Structure of a sliding clamp on DNA. - Cell: 2008 Jan 11, 132(1);43-54 [PubMed:18191219] [DOI]