Difference between revisions of "IS Families/IS1380 family"
| Line 1: | Line 1: | ||
====Original Identification and Distribution==== | ====Original Identification and Distribution==== | ||
| − | The founder member of the family, IS''1380'', was identified in the early 1990s as responsible for the loss of ethanol-oxidizing ability in ''[[wikipedia:Acetobacter|Acetobacter pasteurianus]]'' and is present in a high number of copies<ref name=":0"><pubmed>1657877</pubmed> | + | The founder member of the family, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1380 IS''1380''], was identified in the early 1990s as responsible for the loss of ethanol-oxidizing ability in ''[[wikipedia:Acetobacter|Acetobacter pasteurianus]]'' and is present in a high number of copies<ref name=":0"><pubmed>1657877</pubmed> |
| − | This family is represented by over 150 members from 100 bacterial species. Members of this family ([[:File:Fig. IS1380.1.png|Fig.IS1380.1]]) | + | </nowiki></ref>. In the same year, a second member of the family, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS942 IS''942''], was identified upstream of a [[wikipedia:Beta-lactamase|beta-lactamase]], ''[https://card.mcmaster.ca/ontology/36717 ccrA]'', gene in ''[[wikipedia:Bacteroides_fragilis|Bacteroides fragilis]]'' TAL3636<ref name=":1"><pubmed>1650006</pubmed> |
| − | [[Image:Fig. IS1380.1.png|thumb|center| | + | |
| + | </nowiki></ref> where it was proposed to activate gene expression. | ||
| + | |||
| + | This family is represented by over 150 members from 100 bacterial species. Members of this family ([[:File:Fig. IS1380.1.png|Fig.IS1380.1]]) are between 1665 bp ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1380 IS''1380''] <ref name=":0" />-<ref><nowiki><pubmed>8092854</pubmed></nowiki></ref><ref><nowiki><pubmed>8390818</pubmed></nowiki></ref> and 2071 bp ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISBj1 IS''Bj1'']) in length and each carries a single long open reading frame which includes a potential DDE motif. The terminal '''IRs''' are related and are approximately 15 bp long. They have conserved ends terminating with Cct/c ([[:File:Fig. IS1380.1.png|Fig.IS1380.1]]). | ||
| + | [[Image:Fig. IS1380.1.png|thumb|center|720x720px|'''Fig. IS1380.1.''' The general characteristics of IS''1380'' family. '''Upper panel.''' The average length. '''Bottom panel.''' The common ends identified by [http://weblogo.threeplusone.com/ Weblogo].|alt=]] | ||
====Organization==== | ====Organization==== | ||
| − | Insertion appears to generate a target duplication of 4 bp although several generate 5 bp DRs. Each member exhibits between 30% and 50% amino acid identity in the potential Tpase. Although the majority of their Tpases often include the canonical DDE(6)K/R, several members exhibit other residues in place of the K/R. These include DDE(6)Q or DDE(6)I. A subgroup, IS''942'', composed of 13 members all restricted to [[wikipedia:Bacteroidetes|Bacteroidetes]], include DDE(6)N. None of these differences appear to affect the predicted secondary structure. In addition, to the host-restricted IS''942'' group, two other branches of the Tpase tree are restricted to the [[wikipedia:Actinobacteria|Actinobacteria]]. A single, poorly characterized NCBI database entry ([https://www.ncbi.nlm.nih.gov/protein/WP_018034290.1?report=genpept WP_018034290] and [https://www.ncbi.nlm.nih.gov/genome/annotation_prok/evidence/NF033539/ NF033539]) probably corresponds to an archaeal IS''1380'' member and intriguingly, an orf ([https://www.ncbi.nlm.nih.gov/protein/XP_002337507.1?report=genpept XP_002337507] and [https://www.ncbi.nlm.nih.gov/ipg/XP_002337507.1 XP_002337507.1]) with a 100% match to IS''Lsp5'' (''[[wikipedia:Leptospirillum_ferriphilum|Leptospirillum]]'' sp) has been identified in the genomic sequence of the black cottonwood tree, ''[[wikipedia:Populus_trichocarpa|Populus trichocarpa]]''. Tpases of this family include an insertion domain with a predominantly ↑-strand secondary structure<ref><nowiki><pubmed>20067338</pubmed></nowiki></ref>. | + | Insertion appears to generate a target duplication of 4 bp, although several generate 5 bp DRs. Each member exhibits between 30% and 50% amino acid identity in the potential Tpase. Although the majority of their Tpases often include the canonical DDE(6)K/R, several members exhibit other residues in place of the K/R. These include DDE(6)Q or DDE(6)I. A subgroup, IS''942'', composed of 13 members all restricted to [[wikipedia:Bacteroidetes|Bacteroidetes]], include DDE(6)N. None of these differences appear to affect the predicted secondary structure. In addition, to the host-restricted [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS942 IS''942''] group, two other branches of the Tpase tree are restricted to the [[wikipedia:Actinobacteria|Actinobacteria]]. A single, poorly characterized NCBI database entry ([https://www.ncbi.nlm.nih.gov/protein/WP_018034290.1?report=genpept WP_018034290] and [https://www.ncbi.nlm.nih.gov/genome/annotation_prok/evidence/NF033539/ NF033539]) probably corresponds to an archaeal [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1380 IS''1380''] member and, intriguingly, an orf ([https://www.ncbi.nlm.nih.gov/protein/XP_002337507.1?report=genpept XP_002337507] and [https://www.ncbi.nlm.nih.gov/ipg/XP_002337507.1 XP_002337507.1]) with a 100% match to [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISLsp5 IS''Lsp5''] (''[[wikipedia:Leptospirillum_ferriphilum|Leptospirillum]]'' sp) has been identified in the genomic sequence of the black cottonwood tree, ''[[wikipedia:Populus_trichocarpa|Populus trichocarpa]]''. Tpases of this family include an insertion domain with a predominantly ↑-strand secondary structure<ref><nowiki><pubmed>20067338</pubmed></nowiki></ref>. |
| + | |||
| + | For [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISBj1 IS''Bj1''] a simple phase change around co-ordinate 1365 results in the extension of the potential Tpase orf from 328 to 454 amino acids (similar to that of the other group members) and significantly increases the quality of the alignment. In addition, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISBj1 IS''Bj1''] also carries a 112 bp duplication in its right end. | ||
| − | + | [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1380 IS''1380''] itself <ref name=":0" /> is present in high copy number in the ''[[wikipedia:Acetobacter|Acetobacter pasteurianus]]'' NCI1380 genome and in several strains of acetic acid bacteria. The family contains at least two tIS present in a single copy: [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISMsm12 IS''Msm12''] (''[[wikipedia:Mycobacterium_smegmatis|Mycobacterium smegmatis]]'' ; ''[[wikipedia:TetR|tetR]]'' + [[wikipedia:Methyltransferase|Methyltransferase]]) and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISRop1 IS''Rop1''] (''[[wikipedia:Rhodococcus_opacus|Rhodococcus opacus]]'' ; [[wikipedia:Reverse_transcriptase|reverse transcriptase]]). | |
| − | IS | + | This IS family is distantly related to the eukaryotic PiggyBac TE family (see <ref name=":2"><pubmed>26104701</pubmed> |
| − | + | </nowiki></ref>) which also includes an insertion domain largely in the form of β-strand<ref><nowiki><pubmed>18076328</pubmed></nowiki></ref>. | |
====Activation of Neighboring Genes and Transposition==== | ====Activation of Neighboring Genes and Transposition==== | ||
| − | One member of this family, IS''Ecp1'', has proved clinically important in the spread of extended spectrum resistance to [[wikipedia:Beta-lactam|beta-lactams]]. It was originally identified in plasmid plasmid pST01 from an ''[[wikipedia:Escherichia_coli|E. coli]]'' isolate from the UK (P.D. Stapleton, [https://www.ncbi.nlm.nih.gov/nuccore/AJ242809.1/ AJ242809]) upstream of a [[wikipedia:Cephalosporin|cephalosporinase]] gene, ''[https://www.uniprot.org/uniprot/A0A1B1LTY3 blaCMY]'' and several lines of evidence suggested that it was often associated with bla genes. It was subsequently identified upstream of a gene encoding an extended spectrum CTX-M type beta-lactamase, ''blaCTX-M-1'', and its relationship to the ''[[wikipedia:Beta-lactamase|bla]]'' gene was analysed in some detail: although there appeared to be no endogenous ''blaCTX-M-1'' promoter, it was suggested that a promoter in IS''Ecp1'' was capable of driving ''blaCTX-M-1''<ref name=":3"><pubmed>11470367</pubmed> | + | One member of this family, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''], has proved clinically important in the spread of extended spectrum resistance to [[wikipedia:Beta-lactam|beta-lactams]]. It was originally identified in plasmid plasmid pST01 from an ''[[wikipedia:Escherichia_coli|E. coli]]'' isolate from the UK (P.D. Stapleton, [https://www.ncbi.nlm.nih.gov/nuccore/AJ242809.1/ AJ242809]) upstream of a [[wikipedia:Cephalosporin|cephalosporinase]] gene, ''[https://www.uniprot.org/uniprot/A0A1B1LTY3 blaCMY]'' and several lines of evidence suggested that it was often associated with [[wikipedia:Beta-lactamase|''bla'']] genes. It was subsequently identified upstream of a gene encoding an extended spectrum CTX-M type [[wikipedia:Beta-lactamase|beta-lactamase]], ''blaCTX-M-1'', and its relationship to the ''[[wikipedia:Beta-lactamase|bla]]'' gene was analysed in some detail: although there appeared to be no endogenous ''blaCTX-M-1'' promoter, it was suggested that a promoter in [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] was capable of driving ''blaCTX-M-1''<ref name=":3"><pubmed>11470367</pubmed> |
| + | |||
| + | </nowiki></ref> as had been proposed for [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS942 IS''942''] previously<ref name=":1" />. Further study showed this to be true, and primer extension indicated that a promoter was located within [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] towards its right end of the IS<ref><nowiki><pubmed>12936998</pubmed></nowiki></ref>. Other CTX-ML-beta-lactamases (including CTXM-1, -M-2, and -M-9)<ref name=":3" /><ref name=":4"><pubmed>12205064</pubmed> | ||
| + | |||
| + | </nowiki></ref><ref><nowiki><pubmed>14693512</pubmed></nowiki></ref><ref><nowiki><pubmed>11959547</pubmed></nowiki></ref><ref name=":5"><pubmed>11850241</pubmed> | ||
| + | |||
| + | </nowiki></ref><ref><nowiki><pubmed>15135523</pubmed></nowiki></ref><ref name=":6"><pubmed>12007800</pubmed> | ||
| + | |||
| + | </nowiki></ref> driven by upstream insertions of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] have been identified in various clinical isolates of ''E. coli'' and ''[[wikipedia:Proteus_mirabilis|P. mirabilis]]'' isolated in [[wikipedia:Paris|Paris]] <ref name=":6" /> and in [[wikipedia:Enterobacteriaceae|Enterobacteriaceae]] from [[wikipedia:China|China]]<ref name=":5" /> and [[wikipedia:Poland|Poland]]<ref name=":4" />. | ||
Although there appears to be only a single copy of IS''Ecp1'' associated with the ''blaCTX-M'' genes rather than two flanking copies constituting a compound transposon, several lines of evidence show that this single copy is involved in ''blaCTX-M'' mobilization. | Although there appears to be only a single copy of IS''Ecp1'' associated with the ''blaCTX-M'' genes rather than two flanking copies constituting a compound transposon, several lines of evidence show that this single copy is involved in ''blaCTX-M'' mobilization. | ||
| − | In a first series of studies<ref name=":2" /> detailed analysis of the DNA region surrounding the ''blaCTX-M'' gene a natural plasmid, pILT-3 from ''[[wikipedia:Klebsiella_pneumoniae|K. pneumoniae]]'' ILT-3 ([[:File:Fig. IS1380.2.png|Fig.IS1380.2]]) identified a single IS''Ecp1'' copy upstream containing a potential promoter together with a sequence with some resemblance to an IRR (called IRR1) at some distance downstream which might act as a surrogate IR. Moreover, the structure was flanked by a 5 bp direct repeat as expected for members of the IS''1380'' family suggesting that it could be a transposable unit. The transposition capacity of ''blaCTX-M'' was assessed<ref name=":7"><pubmed>15616333</pubmed> | + | In a first series of studies<ref name=":2" /> detailed analysis of the DNA region surrounding the ''blaCTX-M'' gene a natural plasmid, pILT-3 from ''[[wikipedia:Klebsiella_pneumoniae|K. pneumoniae]]'' ILT-3 ([[:File:Fig. IS1380.2.png|Fig.IS1380.2]]) identified a single [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] copy upstream containing a potential promoter together with a sequence with some resemblance to an IRR (called IRR1) at some distance downstream which might act as a surrogate IR. Moreover, the structure was flanked by a 5 bp direct repeat as expected for members of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1380 IS''1380''] family, suggesting that it could be a transposable unit. The transposition capacity of ''blaCTX-M'' was assessed<ref name=":7"><pubmed>15616333</pubmed></nowiki></ref> by cloning the region from pILT-3 into a high copy number plasmid in ''E. coli'' and using a mating out assay<ref name=":8"><pubmed>6281440</pubmed></nowiki></ref> with the conjugative [https://ecoliwiki.org/colipedia/index.php/pOX38 plasmid pOX38]<ref><nowiki><pubmed>6271456</pubmed></nowiki></ref> as a target. Surprisingly a number of different products were isolated, each carrying at the upstream [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] and its associated ''blaCTX-M'' gene but including different ends ([[:File:Fig. IS1380.3.png|Fig.IS1380.3]] '''A''' and '''C''') but these appear to be true transposition events since they generated 5 bp direct target repeats<ref name=":7" /> ([[:File:Fig. IS1380.3.png|Fig.IS1380.3]] '''B'''). This unexpected result suggests that end recognition (at least at the IRR end) is somewhat relaxed because, since the IR sequences show significant variation. |
| − | [[Image:Fig. IS1380.2.png|thumb|center| | + | [[Image:Fig. IS1380.2.png|thumb|center|680x680px|'''Fig. IS1380.2.''' Genetic Environment of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] transposition module in plasmid pILT-3. The different component mobile genetic elements (a class1 integron, a partial copy of [http://tncentral.ncc.unesp.br/cgi-bin/tn_report.pl?id=Tn1721-X61367.1 Tn''1721''] and the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] based transposable element) are shown in the yellow bar. The associated genes and additional inserted IS are shown below (blue, the integron integrase; red, antibiotic resistance genes lilac, transposases). Above are the left ('''IRL''') and right ('''IRR''') terminal inverted repeats of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] together with the surrogate IRR'. The flanking 5bp direct target repeats are also indicated. The outwardly facing [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] promoter which drives ''bla'' CTX expression is included at the bottom of the figure.|alt=]] |
| − | [[Image:Fig. IS1380.3.png|thumb|center| | + | [[Image:Fig. IS1380.3.png|thumb|center|720x720px|'''Fig. IS1380.3.''' [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] transposition modules in plasmid pILT-3. '''A)''' The relative use of a number of different neighboring surrogate IRR sequences in [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 ISEcp1]-driven transposition of bla CTX is shown together with their DNA sequences, the number of times each was observed and the length of the transposed DNA segments. '''B)''' different target repeat sequences generated by insertion. '''C)''' a diagram of the position of the surrogate '''IRs''' downstream of ''bla'' CTX. |alt=]] |
| + | |||
| + | In an additional set of experiments, it was subsequently shown that [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1B IS''Ecp1B''] can mobilize a [[wikipedia:Beta-lactamase|beta-lactamase]] gene from the chromosome of ''[[wikipedia:Klebsiella|Klebsiella ascorbata]]'' to a plasmid in ''E.coli'' <ref name=":9"><pubmed>28416554</pubmed></nowiki></ref>. The study involved a number of steps. A plasmid carrying an [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] copy was first transformed into a ''[[wikipedia:Klebsiella|K. ascorbata]]'' strain which carries a chromosomal ''blaCTX-M'' gene specifying low level resistance to the [[wikipedia:Cephalosporin|cephalosporin]] [[wikipedia:Cefotaxime|cefotaxime]] (MIC 0.06 micrograms/ml). High level [[wikipedia:Cefotaxime|cefotaxime]] resistance colonies (0.5 or 2 microgram/ml) were scored after growth in liquid culture and occurred at 10-7 per viable cell. They were assumed to represent [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] insertions. Mobilization of ''blaCTX-M'' by [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] was then assessed by a direct mating out assay<ref name=":8" /> from a ''[[wikipedia:Klebsiella|Klebsiella ascorbata]]'' derivative to ''E.coli'' using the conjugative plasmid pOX38Gm<ref name=":9" />. | ||
| + | |||
| + | More recently, to study the increasing number of the relatively high copy number ColE1-type plasmids which are now beginning to be identified having acquired antibiotic resistance genes, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1'']-mediated transposition has been monitored from the chromosome of clinical ''E. coli'' isolates<ref name=":10"><pubmed>30087569</pubmed></nowiki></ref>. A number of these were observed to carry a collection of plasmids including ColE1 derivatives. None of the strains were able to transfer the resistances by conjugation, but three could transfer [[wikipedia:Cefoxitin|cefoxitin]] resistance and the ''blaCMY-2'' gene to a suitable ''E. coli'' recipient at very low frequencies (10-8 –10-9) when plasmid DNA was introduced into a suitable recipient by electroporation. | ||
| + | |||
| + | In these cases, resistance levels were significantly higher than in the original host. The second round of electroporation of plasmid DNA isolated from resistant colonies gave high frequencies of resistance resulting from an insertion from the chromosome into the endogenous plasmid pSC137. Sequence analysis of several plasmid derivatives revealed different insertion events using the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISECP1 IS''ECP1''] '''IRL''' and a surrogate downstream end, all flanked by a 5 bp direct target duplication<ref name=":10" />. The association of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] with [[wikipedia:Beta-lactamase|beta-lactamases]] in clinical isolated appears to be a frequent events<ref name=":11"><pubmed>19596889</pubmed> | ||
| + | |||
| + | </nowiki></ref>. The association of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''] with [[wikipedia:Beta-lactamase|beta-lactamases]] in clinical isolated appears to be a frequent events<ref name=":11" />. This is not limited to antibiotic resistance genes. | ||
| − | + | Although little is known about most other [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1380 IS''1380''] family members, the ability to enhance expression of downstream genes may be a general characteristic. It was previously shown that [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1247 IS''1247''] can activate gene expression and appears to participate in a similar type of transposition as [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1'']<ref><nowiki><pubmed>7868610</pubmed></nowiki></ref>. In ''[[wikipedia:Xanthobacter_autotrophicus|Xanthobacter autotrophicus]]'' GJ10 it can increase resistance to [[wikipedia:Ethyl_bromoacetate|bromoacetate]] by insertion upstream and activation of the [[wikipedia:Haloacetate_dehalogenase|haloacetate dehalogenase]] gene and mobilizes the gene by transposition using a surrogate sequence resembling an IR downstream. More recently, another IS''1380'' family member, IS''Pme1'' from ''[[wikipedia:Paracoccus|Paracoccus methylutens]]''<ref name=":12"><pubmed>18296518</pubmed> | |
| − | + | </nowiki></ref>, which also uses a surrogate downstream IR-like sequence, has been identified. Examples are shown in ([[:File:Fig. IS1380.4.png|Fig.IS1380.4]] '''A''' and '''B'''). These elements have been named TMO (ISX) for Transposition Modules where ISX is the name of the IS involved. Like [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISEcp1 IS''Ecp1''], there is an internal promoter downstream of the transposase gene which is oriented downstream<ref name=":12" />. In addition, another ''[[wikipedia:Paracoccus|Paracoccus]]'' [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1380 IS''1380''] family member, an [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1247 IS''1247''] derivative from ''[[wikipedia:Paracoccus|Paracoccus marcusii]]'' was identified as part of a mosaic transposable element. It appears that a TMO(IS1247a) ([[:File:Fig. IS1380.4.png|Fig.IS1380.4]] '''C''') had inserted into a member of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAs1 IS''As1''] IS family, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPmar4 IS''Pmar4'']<ref name=":13"><pubmed>19187199</pubmed> | |
| − | + | </nowiki></ref>. TMO(IS''1247a'') has flanking 5 bp DR in this transposition unit. It was suggested that the internal outwardly facing [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1247 IS''1247''] promoter could activate the downstream putative [[wikipedia:Alpha/beta_hydrolase_superfamily|alpha/beta hydrolase]] gene<ref name=":13" />. | |
| − | [[Image:Fig. IS1380.4.png|thumb|center| | + | [[Image:Fig. IS1380.4.png|thumb|center|720x720px|'''Fig. IS1380.4.''' '''Transposition modules from ''[[wikipedia:Paracoccus|Paracoccus]]''.''' Three examples of so-called “transposition-modules” from ''[[wikipedia:Paracoccus|Paracoccus marcusii]]'' (Bartosik et al., 2008). There is an internal promoter downstream of the transposase. These elements may use surrogate ends. |alt=]] |
| − | Although no details of the IS''1380'' transposition mechanism are known, these observations suggest that, like other, IS such as IS''91'' (and potentially the IS''CR'' although no formal proof has yet been presented) are capable of “capturing” and driving transposition of neighboring, downstream genes. | + | Although no details of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1380 IS''1380''] transposition mechanism are known, these observations suggest that, like other, IS such as [[IS Families/IS91-ISCR families|IS''91'']] (and potentially the [[IS Families/IS91-ISCR families|IS''CR'']], although no formal proof has yet been presented) are capable of “capturing” and driving transposition of neighboring, downstream genes. |
==Bibliography== | ==Bibliography== | ||
<references /> | <references /> | ||
Revision as of 22:56, 11 August 2021
Contents
Original Identification and Distribution
The founder member of the family, IS1380, was identified in the early 1990s as responsible for the loss of ethanol-oxidizing ability in Acetobacter pasteurianus and is present in a high number of copies[1]. In the same year, a second member of the family, IS942, was identified upstream of a beta-lactamase, ccrA, gene in Bacteroides fragilis TAL3636[2] where it was proposed to activate gene expression.
This family is represented by over 150 members from 100 bacterial species. Members of this family (Fig.IS1380.1) are between 1665 bp (IS1380 [1]-[3][4] and 2071 bp (ISBj1) in length and each carries a single long open reading frame which includes a potential DDE motif. The terminal IRs are related and are approximately 15 bp long. They have conserved ends terminating with Cct/c (Fig.IS1380.1).
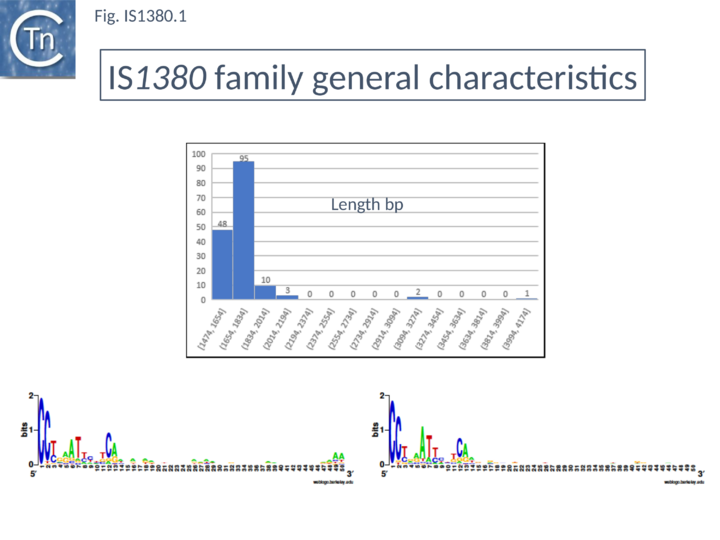
Organization
Insertion appears to generate a target duplication of 4 bp, although several generate 5 bp DRs. Each member exhibits between 30% and 50% amino acid identity in the potential Tpase. Although the majority of their Tpases often include the canonical DDE(6)K/R, several members exhibit other residues in place of the K/R. These include DDE(6)Q or DDE(6)I. A subgroup, IS942, composed of 13 members all restricted to Bacteroidetes, include DDE(6)N. None of these differences appear to affect the predicted secondary structure. In addition, to the host-restricted IS942 group, two other branches of the Tpase tree are restricted to the Actinobacteria. A single, poorly characterized NCBI database entry (WP_018034290 and NF033539) probably corresponds to an archaeal IS1380 member and, intriguingly, an orf (XP_002337507 and XP_002337507.1) with a 100% match to ISLsp5 (Leptospirillum sp) has been identified in the genomic sequence of the black cottonwood tree, Populus trichocarpa. Tpases of this family include an insertion domain with a predominantly ↑-strand secondary structure[5].
For ISBj1 a simple phase change around co-ordinate 1365 results in the extension of the potential Tpase orf from 328 to 454 amino acids (similar to that of the other group members) and significantly increases the quality of the alignment. In addition, ISBj1 also carries a 112 bp duplication in its right end.
IS1380 itself [1] is present in high copy number in the Acetobacter pasteurianus NCI1380 genome and in several strains of acetic acid bacteria. The family contains at least two tIS present in a single copy: ISMsm12 (Mycobacterium smegmatis ; tetR + Methyltransferase) and ISRop1 (Rhodococcus opacus ; reverse transcriptase).
This IS family is distantly related to the eukaryotic PiggyBac TE family (see [6]) which also includes an insertion domain largely in the form of β-strand[7].
Activation of Neighboring Genes and Transposition
One member of this family, ISEcp1, has proved clinically important in the spread of extended spectrum resistance to beta-lactams. It was originally identified in plasmid plasmid pST01 from an E. coli isolate from the UK (P.D. Stapleton, AJ242809) upstream of a cephalosporinase gene, blaCMY and several lines of evidence suggested that it was often associated with bla genes. It was subsequently identified upstream of a gene encoding an extended spectrum CTX-M type beta-lactamase, blaCTX-M-1, and its relationship to the bla gene was analysed in some detail: although there appeared to be no endogenous blaCTX-M-1 promoter, it was suggested that a promoter in ISEcp1 was capable of driving blaCTX-M-1[8] as had been proposed for IS942 previously[2]. Further study showed this to be true, and primer extension indicated that a promoter was located within ISEcp1 towards its right end of the IS[9]. Other CTX-ML-beta-lactamases (including CTXM-1, -M-2, and -M-9)[8][10][11][12][13][14][15] driven by upstream insertions of ISEcp1 have been identified in various clinical isolates of E. coli and P. mirabilis isolated in Paris [15] and in Enterobacteriaceae from China[13] and Poland[10].
Although there appears to be only a single copy of ISEcp1 associated with the blaCTX-M genes rather than two flanking copies constituting a compound transposon, several lines of evidence show that this single copy is involved in blaCTX-M mobilization.
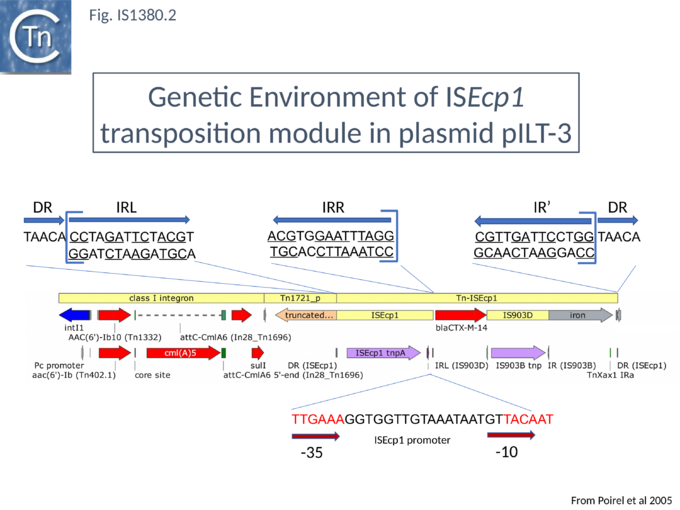
In a first series of studies[6] detailed analysis of the DNA region surrounding the blaCTX-M gene a natural plasmid, pILT-3 from K. pneumoniae ILT-3 (Fig.IS1380.2) identified a single ISEcp1 copy upstream containing a potential promoter together with a sequence with some resemblance to an IRR (called IRR1) at some distance downstream which might act as a surrogate IR. Moreover, the structure was flanked by a 5 bp direct repeat as expected for members of the IS1380 family, suggesting that it could be a transposable unit. The transposition capacity of blaCTX-M was assessed[16] by cloning the region from pILT-3 into a high copy number plasmid in E. coli and using a mating out assay[17] with the conjugative plasmid pOX38[18] as a target. Surprisingly a number of different products were isolated, each carrying at the upstream ISEcp1 and its associated blaCTX-M gene but including different ends (Fig.IS1380.3 A and C) but these appear to be true transposition events since they generated 5 bp direct target repeats[16] (Fig.IS1380.3 B). This unexpected result suggests that end recognition (at least at the IRR end) is somewhat relaxed because, since the IR sequences show significant variation.
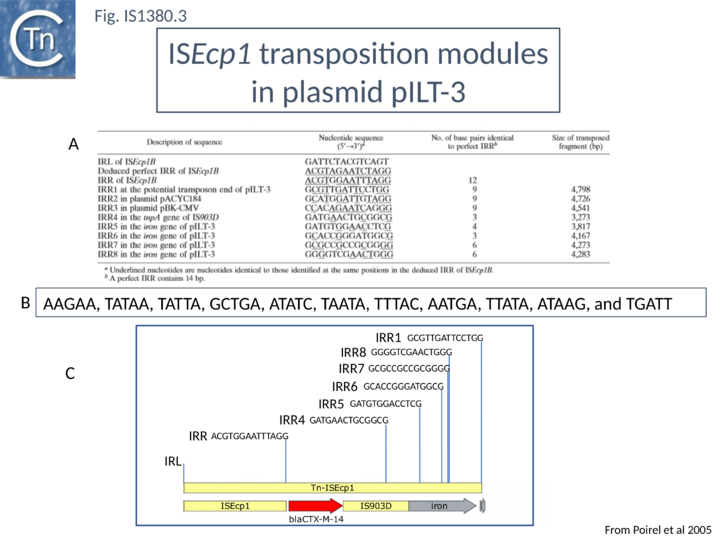
In an additional set of experiments, it was subsequently shown that ISEcp1B can mobilize a beta-lactamase gene from the chromosome of Klebsiella ascorbata to a plasmid in E.coli [19]. The study involved a number of steps. A plasmid carrying an ISEcp1 copy was first transformed into a K. ascorbata strain which carries a chromosomal blaCTX-M gene specifying low level resistance to the cephalosporin cefotaxime (MIC 0.06 micrograms/ml). High level cefotaxime resistance colonies (0.5 or 2 microgram/ml) were scored after growth in liquid culture and occurred at 10-7 per viable cell. They were assumed to represent ISEcp1 insertions. Mobilization of blaCTX-M by ISEcp1 was then assessed by a direct mating out assay[17] from a Klebsiella ascorbata derivative to E.coli using the conjugative plasmid pOX38Gm[19].
More recently, to study the increasing number of the relatively high copy number ColE1-type plasmids which are now beginning to be identified having acquired antibiotic resistance genes, ISEcp1-mediated transposition has been monitored from the chromosome of clinical E. coli isolates[20]. A number of these were observed to carry a collection of plasmids including ColE1 derivatives. None of the strains were able to transfer the resistances by conjugation, but three could transfer cefoxitin resistance and the blaCMY-2 gene to a suitable E. coli recipient at very low frequencies (10-8 –10-9) when plasmid DNA was introduced into a suitable recipient by electroporation.
In these cases, resistance levels were significantly higher than in the original host. The second round of electroporation of plasmid DNA isolated from resistant colonies gave high frequencies of resistance resulting from an insertion from the chromosome into the endogenous plasmid pSC137. Sequence analysis of several plasmid derivatives revealed different insertion events using the ISECP1 IRL and a surrogate downstream end, all flanked by a 5 bp direct target duplication[20]. The association of ISEcp1 with beta-lactamases in clinical isolated appears to be a frequent events[21]. The association of ISEcp1 with beta-lactamases in clinical isolated appears to be a frequent events[21]. This is not limited to antibiotic resistance genes.
Although little is known about most other IS1380 family members, the ability to enhance expression of downstream genes may be a general characteristic. It was previously shown that IS1247 can activate gene expression and appears to participate in a similar type of transposition as ISEcp1[22]. In Xanthobacter autotrophicus GJ10 it can increase resistance to bromoacetate by insertion upstream and activation of the haloacetate dehalogenase gene and mobilizes the gene by transposition using a surrogate sequence resembling an IR downstream. More recently, another IS1380 family member, ISPme1 from Paracoccus methylutens[23], which also uses a surrogate downstream IR-like sequence, has been identified. Examples are shown in (Fig.IS1380.4 A and B). These elements have been named TMO (ISX) for Transposition Modules where ISX is the name of the IS involved. Like ISEcp1, there is an internal promoter downstream of the transposase gene which is oriented downstream[23]. In addition, another Paracoccus IS1380 family member, an IS1247 derivative from Paracoccus marcusii was identified as part of a mosaic transposable element. It appears that a TMO(IS1247a) (Fig.IS1380.4 C) had inserted into a member of the ISAs1 IS family, ISPmar4[24]. TMO(IS1247a) has flanking 5 bp DR in this transposition unit. It was suggested that the internal outwardly facing IS1247 promoter could activate the downstream putative alpha/beta hydrolase gene[24].
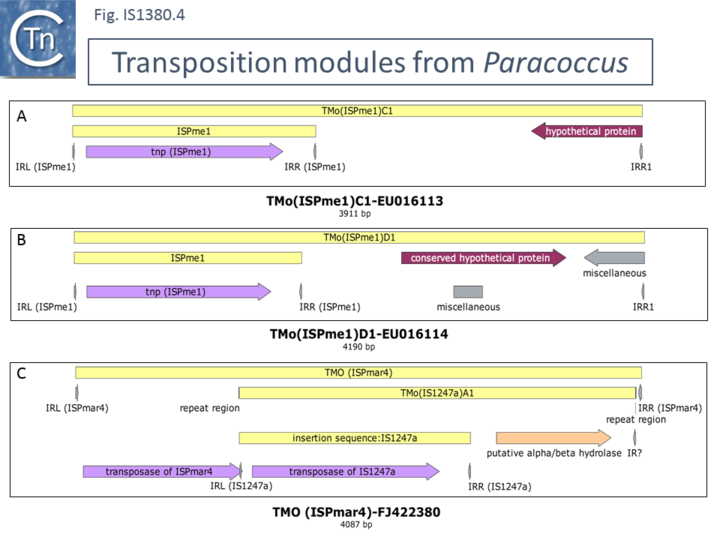
Although no details of the IS1380 transposition mechanism are known, these observations suggest that, like other, IS such as IS91 (and potentially the ISCR, although no formal proof has yet been presented) are capable of “capturing” and driving transposition of neighboring, downstream genes.
Bibliography
- ↑ 1.0 1.1 1.2 Takemura H, Horinouchi S, Beppu T . Novel insertion sequence IS1380 from Acetobacter pasteurianus is involved in loss of ethanol-oxidizing ability. - J Bacteriol: 1991 Nov, 173(22);7070-6 [PubMed:1657877] [DOI] </nowiki>
- ↑ 2.0 2.1 Rasmussen BA, Kovacs E . Identification and DNA sequence of a new Bacteroides fragilis insertion sequence-like element. - Plasmid: 1991 Mar, 25(2);141-4 [PubMed:1650006] [DOI] </nowiki>
- ↑ <pubmed>8092854</pubmed>
- ↑ <pubmed>8390818</pubmed>
- ↑ <pubmed>20067338</pubmed>
- ↑ 6.0 6.1 </nowiki>
- ↑ <pubmed>18076328</pubmed>
- ↑ 8.0 8.1 </nowiki>
- ↑ <pubmed>12936998</pubmed>
- ↑ 10.0 10.1 Baraniak A, Fiett J, Hryniewicz W, Nordmann P, Gniadkowski M . Ceftazidime-hydrolysing CTX-M-15 extended-spectrum beta-lactamase (ESBL) in Poland. - J Antimicrob Chemother: 2002 Sep, 50(3);393-6 [PubMed:12205064] [DOI] </nowiki>
- ↑ <pubmed>14693512</pubmed>
- ↑ <pubmed>11959547</pubmed>
- ↑ 13.0 13.1 </nowiki>
- ↑ <pubmed>15135523</pubmed>
- ↑ 15.0 15.1 </nowiki>
- ↑ 16.0 16.1 </nowiki>
- ↑ 17.0 17.1 </nowiki>
- ↑ <pubmed>6271456</pubmed>
- ↑ 19.0 19.1 </nowiki>
- ↑ 20.0 20.1 </nowiki>
- ↑ 21.0 21.1 </nowiki>
- ↑ <pubmed>7868610</pubmed>
- ↑ 23.0 23.1 Bartosik D, Putyrski M, Dziewit L, Malewska E, Szymanik M, Jagiello E, Lukasik J, Baj J . Transposable modules generated by a single copy of insertion sequence ISPme1 and their influence on structure and evolution of natural plasmids of Paracoccus methylutens DM12. - J Bacteriol: 2008 May, 190(9);3306-13 [PubMed:18296518] [DOI] </nowiki>
- ↑ 24.0 24.1 </nowiki>