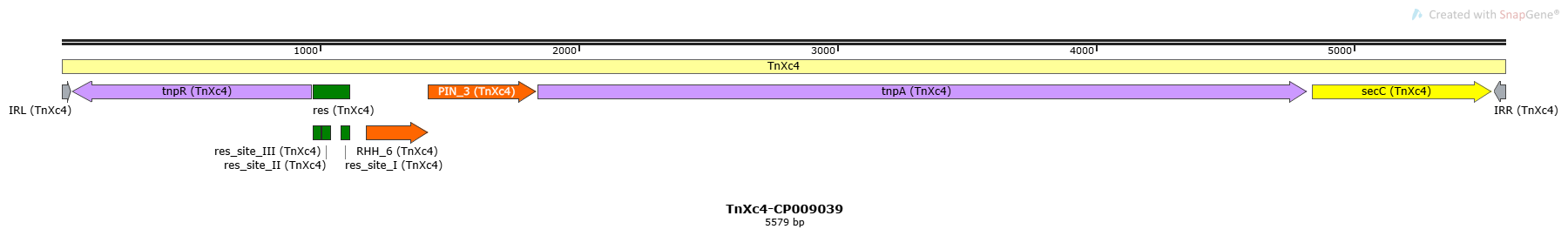
Mobile element type:
Transposon
Name:
TnXc4
Synonyms:
Tn7210
Accession:
TnXc4-CP009039
Family:
Tn3
Group:
Tn3
First isolate:
Yes
Partial:
ND
Evidence of transposition:
ND
Host Organism:
Xanthomonas citri subsp. citri strain AW16
Date of Isolation:
2015
Country:
U.s.a.
Molecular Source:
plasmid pXCAW58
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-38 | + | 38 | GGGGTTCGGG GAGCAATGGA ACAGGGAAGT CAGTTAAG |
| IRR | 5542-5579 | - | 38 | GGGGTTCGGG GAGCAATGGA ACAGGGAAGT CAGTTAAG |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res | TnXc4 | 973-1112 | + | 140 | TTTTGCTTTA CTCGTTAATT TTTAATAATA TCAAACTTTG ATTGTACAAA CCACTATTAC AGACTGCTTG TTGGCTTACG GACACCTCGC GCCGATCGCC CCATCAGAGT TCATAAAAAC GATCGTTTTA TTGAACCGTT |
| res_site_III | TnXc4 | 973-1001 | + | 29 | TTTTGCTTTA CTCGTTAATT TTTAATAAT |
| res_site_II | TnXc4 | 1005-1040 | + | 36 | AAACTTTGAT TGTACAAACC ACTATTACAG ACTGCT |
| res_site_I | TnXc4 | 1080-1112 | + | 33 | AGTTCATAAA AACGATCGTT TTATTGAACC GTT |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| tnpR | TnXc4 | 42-965 | - | Accessory Gene | Resolvase |
| RHH_6 | TnXc4 | 1180-1419 | + | Passenger Gene | Antitoxin |
| PIN_3 | TnXc4 | 1419-1838 | + | Passenger Gene | Toxin |
| tnpA | TnXc4 | 1843-4818 | + | Transposase | |
| secC | TnXc4 | 4837-5535 | + | Passenger Gene | Plant Pathogenicity |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | TnXc4 | 307 | 42-965 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MKIGYARVST REQNPALQVD SLKAAGCERI YQDVASGAKT ARPALDELLG QLRGGDVLVI WKLDRMGRSL KHLVELVGSL MERKVGLLSL NDPIDTTSAQ GRFVFNLFAT LAEFERELIR ERTQAGLTAA RARGRVGGRP KGLSPQAEAT ALAAETLYRE RKLSVAAIAQ KLHLSKSTLY SYLRHRGVEI GPYKQSAQSP INVSVVESGN ADQPKVATIL LTLRIENNSK FVRGKKRSIE HVEFFDLAQY DATRRPNGEY ELKVPYDTDE ELDEAVNDLL TDIACGADDR HCFSESHARM EGTDRHW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | RHH_6 | RHH_6 | TnXc4 | 79 | 1180-1419 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antitoxin | ||||||||||
| Sequence Family: | RHH_6 (Pfam:PF16762) | ||||||||||
| Protein Sequence: | MNTVRWNIAV SPDVDQSVRM FIAAQGGGRK GDLSRFIEDA VRAYLFERAV EQAKAATVGM GETELNDLID EAVQWAREH |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | PIN_3 | PIN_3 | TnXc4 | 139 | 1419-1838 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Toxin | ||||||||||
| Sequence Family: | PIN_3 (Pfam:PF13470) | ||||||||||
| Target: | single stranded RNA | ||||||||||
| Comment: | tRNA(fMet)-specific endonuclease | ||||||||||
| Protein Sequence: | MRVVLDTNVL LAALISSHSP PDIIYRGWLA ARFELVTGTA QLDELRRVSR YPKIKAILPA HRVGTMINNM QRAVVLHVLP PLPDRIEVND PNDAFLLAMA LASEADYLVT GDRRAGLLQR GSIGRTRIVT PVTFCAEAL |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | TnXc4 | 991 | 1843-4818 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MPVSFLSTTQ RERYGRYPDT LSSEELARYF HLDDDDREWI ATKRRDSSRL GYALQLTTAR FLGTFLEDPT AVPSPVLHTL SSQLGIADPS DCVIDYRTTR QRWQHTSEIR TRYGYREFTG TGVQFRLGRW LCALCWTGTD RPSALFDYAN GWLVGHKVLL PGVTLLERFI AEIRSRMESR LWRLLVHGVT PEQRQRLDDL LKLVEGSRQS WLDRLRKGPV RVSAPALVAA LLRIETVRGL GIKLPGTHVP PSRIAALARF ASTAKVSAVA RLPEVRRIAT LVAFVHCLEA SAQDDAIDVL DLLLRELFTK AEKEDRKVRQ RSLKDLDRAA STLAEACRML LDPALPDGEL RERVYAAIGH DELAQALNEV RGLVRPPNDV FYTELEARKA TVSRFLPALL RVIRFDANPA AQPLAQALQW LHEKPDHDPP TAIVGKAWQR HVVQDDGRIN ATAYSFCALD KLRSAIRRRD VFISPSWRYA DPRAGLLAGA EWEASRPIVC RSLSLSAQPE ATLSELTREL DETYRRVAAR LPQNDAVRFE NVGDKTELVL SPLEALEEPP SLIALRNEIK ARMPRVDLPE ILLEVAGRTG CMEAFTHLTE RTARAADLTT SLCAVLMAEA CNTGPEPLVR PDTPALKRDR LMWVDQNYVR DDTLTACNAV LVAAQSRIAL ARTWGGGDVA SADGMRFVVP VRTIHAGPNP KYFNRGRGVT WYNLLSDQRT GLNAITVPGT LRDSLILLAV VLEQQTELQP TQIMTDTGAY SDLVFGLFRL SNYRFCPRLA DVGGTRFWRV DPDADYGDLN ALARQRVNLD RITPHWDDVL RLVGSLKLGL VPAMGIMRTL QVDERPTSLA QAIAEIGRID KTIHTLNFID DEARRRATLL QLNLGEGRHS LAREVFHGKR GELFQRYREG QEDQLSALGL VVNMIVLWNT LYMDAVLTQL RSEGYPVKPE DEARLSPFGH EHINMLGRYS FSVPEAVARG ELRPLTKPND P |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | secC | SecC | TnXc4 | 232 | 4837-5535 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Plant Pathogenicity | ||||||||||
| Comment: | contains 2 target sites for ISXac1 insertions | ||||||||||
| Protein Sequence: | MQQMTQQPLN DAQLDRLGDF LEGVGAPAMN LEMLDGFFAA LICGPETVLP SEYLPQVFGE DHCFDSNDQA AEILGLVMRH WNTIASELFR TLEKDDVYLP VLLEDADGAV HGNDWARGFM RGIQLRPNSW QELIGSEEFG GPMLPIMILT HEHDPDPAMR PPEIAPDKRD ELLQSLVAGL THIYRYFASH RQLATQGPLR RQGPKIGRND QCPCGSGRKY KHCCATSAPT FH |
||||||||||
1. Zhang Y, Jalan N, Zhou X, Goss E, Jones JB, Setubal JC, Deng X, Wang N.
Positive selection is the main driving force for evolution of citrus canker-causing Xanthomonas.. ISME J. 2015 Oct;9(10):2128-38. doi: 10.1038/ismej.2015.15. Epub 2015 Feb 17. PubMed ID:
25689023
.