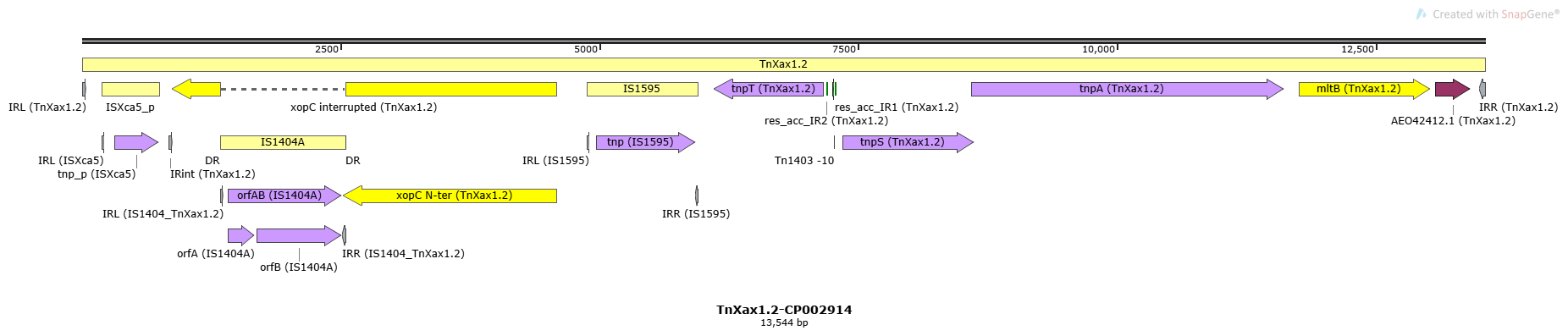
Mobile element type:
Transposon
Name:
TnXax1.2
Synonyms:
Tn7208
Accession:
TnXax1.2-CP002914
Family:
Tn3
Group:
Tn4651
First isolate:
Yes
Partial:
ND
Evidence of transposition:
ND
Host Organism:
Xanthomonas axonopodis pv. citrumelo F1
Date of Isolation:
2011
Country:
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-46 | + | 46 | GAGGGTCGGC AGGGATTCGT GTAAAAGACA GCCAAAAGTG AGCTAA |
| IRint | 838-876 | + | 39 | GGGGCATCAC GATTGTTTCG AGCCTCTGCC CAACTAAAG |
| IRR | 13500-13544 | - | 45 | GAGGGTCGAC AGGGATTTGT GTAAAAAACA GCCAAAATTG AGCTA |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res_acc_IR2 | TnXax1.2 | 7197-7205 | + | 9 | TACCATGTA |
| res_acc_IR2 | TnXax1.2 | 7254-7267 | + | 14 | TCACGCGGGA TAAT |
| res_acc_IR1 | TnXax1.2 | 7273-7286 | - | 14 | TAAGGCGGGA TAAT |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| orfAB | IS1404A | 1410-2512 | + | Transposase | |
| orfA | IS1404A | 1410-1673 | + | Accessory Gene | Regulator |
| orfB | IS1404A | 1691-2512 | + | Transposase | |
| xopC N-ter | TnXax1.2 | 2522-4582 | - | Passenger Gene | Plant Pathogenicity |
| tnp | IS1595 | 4972-5934 | + | Transposase | |
| tnpT | TnXax1.2 | 6109-7164 | - | Accessory Gene | Resolvase |
| tnpS | TnXax1.2 | 7348-8622 | + | Accessory Gene | Resolvase |
| tnpA | TnXax1.2 | 8594-11620 | + | Transposase | |
| mltB | TnXax1.2 | 11755-13032 | + | Passenger Gene | Plant Pathogenicity |
| AEO42412.1 | TnXax1.2 | 13075-13419 | + | Passenger Gene | Hypothetical |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | orfAB | OrfAB | IS1404A | 367 | 1410-2512 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | produced by -1 translational frameshifting between orfA and orfB | ||||||||||
| Protein Sequence: | VKKRFSEEQI IGFLREADAG VAIKDLCRRH GFSEASYYLW RSKFGGMSVP DAKRLKDLES ENARLKKLLA EQLFENDLIK DALRKKW*AH RRVACWCASG SGVVPASVVR WQ*SA*APVR CDIARAKIAT SNCANASVRW RIAIAVMVWG *SISNYDRKD AS*TTSVWSG CTASSSCRSG AASAKRC**E SVSRCCGHRR PIRCGRWISC STAAPKAE*S SVW*SWTMQP TKRSPSRWSA QSRAMA*HAC WIGWRTVAAC HR*SAQITAR SFAAKRWWPG HMRVACSYG* SSLASQTRTP TSNRSMAGCA TNASTSTGSQ PCCMRVPRSN AGDANTTRSD PRRPLAA*RH LRMPNSWPTP ISSTPDS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | orfA | OrfA | IS1404A | 87 | 1410-1673 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Regulator | ||||||||||
| Comment: | regulatory protein | ||||||||||
| Protein Sequence: | MKKRFSEEQI IGFLREADAG VAIKDLCRRH GFSEASYYLW RSKFGGMSVP DAKRLKDLES ENARLKKLLA EQLFENDLIK DALRKKW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | orfB | OrfB | IS1404A | 273 | 1691-2512 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MLVREWIGRG ASERRALAVI GMSASALRYC PREDRNVELR ERICALAHRH RRYGVGMIYL KLRQEGRIVN YKRVERLYRE QQLQVRRRKR KKVLIGERQP LLRPSQANQV WSMDFVFDRS AEGRVIKCLV IVDDATHEAI AIEVERAISG HGVTRVLDRL AHSRGLPQVI RTDNGKEFCG KAMVAWAHAR GVQLRLIQPG KPNQNAYVES FNGRLRDECL NEHWFPTLLH ARTEIERWRR EYNEERPKKA IGGMTPSAYA QQLANTDIIN PGL |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | xopC N-ter | XopC N-ter | TnXax1.2 | 686 | 2522-4582 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Plant Pathogenicity | ||||||||||
| Protein Sequence: | MKTSSASKSH YTGQFPQGDL QPKTPIPVGR NSEIPKDAAL SAQLKGLRPL DRKPSKMPTS SSDLQAVNKM IAENRSNDVG KGQFLLSTQS LGSAERVDSQ NYHSRNESGK APLFFSEFAT EKPSLQSVKS LSQGKQDYYA VRHVGKRGRD LFTDEPIEGE NSKLGRLKTS PLLSTQSQGT RAIRAFAATA TIDKSRGEYV ARLHRHVVEA LGDEGGVVHL LRPTRDHYAE DSTLNFFTFC EQTELASSLN SLEAISAKKE DRKPIVFTDH KSLVGLSRTS RTEASPIGRL VQQPYFVTSD LKRGDKVVIT DDHIQAGGSM LAMEAAAKEA GVDVLALATL SAHPFSPQLT MSSEVSAFLD ETLAAWDPQG KVTERLAAIG MPREKLTNSE AMILIAYAID PASDSAILRF ESIQNNLFKR AHLHNVGIDP DAPDTLEQMQ KLSSMQEMAG TDSARHFVDD AKVLEGEHDS LIPILKMKPA SPDEIVKELD QVSLASRSSI QASEVKQVVV LDWDDCLRDE KGLNYKLMHN ALAITAREHA STLPELGDAV AVLHSKLNDA EPVDEEAPLL MKNQKDFSSY LMGRQSIYKR HIIEDFVRKM LPGISEHKAA SINNAVYSNF VREYKMLVKP EVSKKNYSRD VPFPDIELSL LPGAKEILEK SRNANSRVIL ISNRGHGDLA TSPRSE |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnp | Tnp | IS1595 | 320 | 4972-5934 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MSINAVQFQA GLSMPEFFAF YGTEAKCYRA LYTWRWPQGF RCPVCAGRVR SRFKRRAAIY YQCCACRHQT SLMAGTMFEG TKLPLRTWML ALHLLTSTKT NMAALELMRH LGVNYKTAWR MKHKIMQVMA EREATRKLAG FVQIDDAYLG GERNGGKAGR GSENKQAFLI AVQTDATFTA PRFVVIEPVR SFDNTSLQDW IARRLAPECE VYTDGLACFR RLEDAGHAHT TLDTGGGRAA TETAGARWLN VVLGNLKRAI SGVYHAIAQG KYARRYLGEA AYRFNRRFRL REMLPRLATA MMQSNPCPEP VLRAASNFHG |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpT | TnpT | TnXax1.2 | 351 | 6109-7164 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Protein Sequence: | MARSGLYKSD VQRARDRLRA TGTHPSVDAV RVALGNTGSK TTIHRYLREL EEEEGQGVGA KMAVSDALQD LIARLAERLH SEADTVVAQA QARFQAQLQE RTQALEQARH EAGSLMTQLQ RCETALQAER EAGDAARSEV ARRTTELAQL EERIAGLTAR LAEHDAHAKS LEHKHEHARE ALEHYRTSVK DQREQEQRRH EHQVQELQVA LRQANEALTA KNHDLMQLNR ENGQWLERQT RLERELAQAR QRADAQQRER DALRLAAAEH QALQVRWTQD VHALEGVRTE LSAARTELVE ERQRREHAEA DTLRATVRLS TLEQLLAQLR LAHSVGELEN AIAAGAMPAG K |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpS | TnpS | TnXax1.2 | 424 | 7348-8622 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Tyrosine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Tyrosine | ||||||||||
| Protein Sequence: | MSSVKQYLEA ATRANTERAY AGAIRHFEVE WGGHLPATAE QVACYLAAYA GQLALNTLRH RLAALAQWHQ VHGFADPTRA PVVRQVLKGI QTLHPSVEKR ATPLQLTQLG QVTTWLEDAA AAARSRGDRA SELRHLRDRA FLLLGFWRGF RGDELARLQV DHLRLVPGEG MTCFLPHSKG DRRHAGTTYK VPALSRWCPV ASTSSWIAAA ALTDGPVFRA INQWGGVADT ALHPNSLVPL LRRIFGEAGL ASPNTYSGHS LRRGFAGWAN ANGWDVKALM EYVGWRDVHS AMRYLDGADP FARQRIEASL SPATPPLLAL AAPAPDPVPT TAVEATVTLT RFNSRVRGLA KAHRLIEQIC LQPHQAQRLN ADGTRYRLAI AAVDEAAFEE TIAMLLDEMH RIADNHQCFL AVALRDEAGG RHWD |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | TnXax1.2 | 1008 | 8594-11620 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MKRAGAIGTE LGMTTLHETA YPRLKPDPTA KELQDIYTPT AAELQCVRNI ATGPATRLAL LLHLKLFQRL GYFTTLIEVP ERIVQHVAQT LGMRRVPADR LAGYDASGAK RGHLAQLRAF LNVCPLDAAG RDWLGTVAET AARTKHIVPD IVNVMLEELV HHRFELPAFS TLERIAIAAR ERVHDAHYRQ IADALSPTMR TLIDNLLLTP PGSHHSDWHT LKREPKRPTN KEVRHYLRHI QRLRILAEQL PPIDVSVPKL KQFRAMARAL DAAELAELVP IKRYALAAIF IRSQYRKTLD DAADLFIRLI QNLENTAQQK LIAYQLEHSK RADALIGQLR EILQAYQVEG TDTERVGAIA GVLVADIALL TAECDEHMAY AGRNYLPFLL APYGTLRPLL FNCLEIMGLR AASQDPSMER MIGAVLALRS QRRETIDAAS LGVATTDLTW LSSAWRKHVM PKALAAASPG WIHRKYFELA VLAQIKDELK SGDLYIPHGE RYDDYREQLV DEATLAQELD AYGEVSGVAT DAADFVQGLR TELTTLADAV DARFSDNLHA SMLDGRLVLK RLQGAQVTQA IATVDSAITD RLPPTSIVDV LVDTTRWLDL HVHFRPIAGT DARVDDLLRR VITTLFCYGC NLGPTQTARS VKGFSRRQIS WLNLKYVTDE TLDKAIVQVI NMYNKFELPG YWGSGKSASA DGTKWNVYEQ NLLSEYHIRY GGYGGIGYYH VSDKYIALFS HFIPCGIHEA VYILDGMLAN RSDIQPDTVH GDTQAQSFPV FGLAHLLGIN LMPRIRNIKD LVFSRPEPGR TYENIQALFG DSIDWTLIET HVHDMLRVAI SIKLGKITAS TILRRLGTYS RKNKLYWAFR ELGKAVRTLF LLRYIDDVEV RKTIHAATNK SEEFNGFVKW AFFGGEGIIA ENVQHEQRKI VRYNQLVANL VILHNVEQMT RVLAELRDEG SNISPEVLAG LSPYRTSHIN RFGDYTLDLK RQVEPIDFSR RILAATTR |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | mltB | MltB | TnXax1.2 | 425 | 11755-13032 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Plant Pathogenicity | ||||||||||
| Target: | cell wall | ||||||||||
| Comment: | Membrane-bound lytic murein transglycosylase B precursor | ||||||||||
| Protein Sequence: | MIMPSRLLRL TLGVSVCVAA TSVAAQAIAP ETTASSATDA GGQQQDPAPT SSFEQWLADF RQRALAAGIG ATTLDNALAG VTPDPAVHEL DQRQPEFTQY LWDYLDARVT PAAIQEGQQL LISQHALFEK LRQHYGVDPG ILTAIWSMES GYGKQIGDFY VIRSLATLAH EGRRTTYGNT QLLAALQILQ TEKSIDRSQL VGSWAGAMGQ TQFIPSTYRD YAVDEDGDQK RDVWNSKADA LGSAANYLKQ NNWTSAVPWG QEVQLSAGFD YAQADLTIKK TVAEWQRLGV APRRPIAPAL AQQLASVLLP TGYRGPAFLV FDNFRSILRY NNSTAYALAV GLLADGYAGR AGVEQPWPKD DPPLNSTAQI TELQQRLTDK GFDVGGIDGV LGAQTRQGIR AFQRSQQLPQ DGYASTSLLA RLRAP |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | AEO42412.1 | AEO42412.1 | TnXax1.2 | 114 | 13075-13419 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Hypothetical | ||||||||||
| Protein Sequence: | MLTNNEALSM LRKSIILAKI SSFLAAIAFG FLFLSISAPS QADDLGAVSV HGDIQPANPT DNQVRSAHQQ QALGKCKQQF VNSSSAYLAS WAVTAGGKHP DYWNINSVWI CSSG |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| ISXca5_p-CP002914 | ISXca5_p | insertion sequence | 196-747 | + | 552 |
| IS1404A-U45994 | IS1404A | insertion sequence | 1342-2544 | + | 1203 |
| IS1595-AF249895 | IS1595 | insertion sequence | 4881-5952 | + | 1072 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRL | IS1595 | 4881-4904 | + | 24 | CCTGATTAGC CCTGACCTTC AGCC |
| IRR | IS1595 | 5929-5952 | - | 24 | CCTGATTAGC CCTGACTCTC AGCC |
1. Jalan N, Aritua V, Kumar D, Yu F, Jones JB, Graham JH, Setubal JC, Wang N.
Comparative genomic analysis of Xanthomonas axonopodis pv. citrumelo F1, which causes citrus bacterial spot disease, and related strains provides insights into virulence and host specificity.. J Bacteriol. 2011 Nov;193(22):6342-57. doi: 10.1128/JB.05777-11. Epub 2011 Sep 9. PubMed ID:
21908674
.