Mobile element type:
Transposon
Name:
TnPfu1
Synonyms:
Tn7220
Accession:
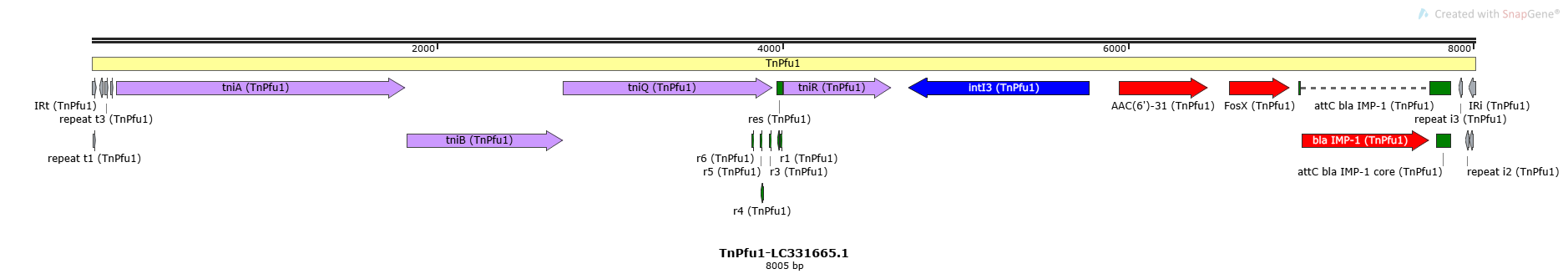
TnPfu1-LC331665.1
Family:
Tn402
Group:
First isolate:
Yes
Partial:
ND
Evidence of transposition:
Yes
Host Organism:
Pseudomonas fulva KUN1351
Date of Isolation:
2018
Country:
Japan
Molecular Source:
chromosome
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRt | 1-33 | + | 33 | TGTCATTTTC AGAAGACGAC TGCACCAATT GAT |
| repeat t1 | 9-27 | + | 19 | TCAGAAGACG ACTGCACCA |
| repeat t2 | 49-67 | - | 19 | TCAGGAGCTG GCTGCACAA |
| repeat t3 | 78-97 | + | 20 | TCAGAAGTGA TCTGCACCAA |
| repeat t4 | 110-128 | + | 19 | TCAATACTCG TGTGCACCA |
| repeat i3 | 7916-7935 | - | 20 | TCATGCAATG GCGGGCAGCA |
| repeat i2 | 7956-7974 | - | 19 | TCAGAAGCCG ACTGCACTA |
| IRi | 7973-8005 | - | 33 | TGTCGTTTTC AGAAGGCGGC TGCACTGAAC GTC |
| repeat i1 | 7978-7997 | - | 20 | TCAGAAGGCG GCTGCACTGA |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| r6 | TnPfu1 | 3824-3837 | + | 14 | TTGGAACGGT TGCG |
| r5 | TnPfu1 | 3870-3883 | + | 14 | AATTTCTGTC ACAT |
| r4 | TnPfu1 | 3875-3888 | - | 14 | AGGTAATGTG ACAG |
| r3 | TnPfu1 | 3926-3939 | + | 14 | GGGTTAAGTG ACAA |
| res | TnPfu1 | 3967-4000 | + | 34 | ACACTGTCAC ATAATCGAAC GTATACGTGA CAGG |
| r2 | TnPfu1 | 3970-3983 | - | 14 | CGATTATGTG ACAG |
| r1 | TnPfu1 | 3986-3999 | + | 14 | CGTATACGTG ACAG |
| attC bla IMP-1 core | TnPfu1 | 7785-7869 | + | 85 | GGACAGTTTG TAAGTTGCGC TTTTGTGGTT TGCTTCGCAA AGTATTCCAC AACGCGCAAC TTACAAACTG CCGCTGAACT TAGCG |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| tniA | TnPfu1 | 142-1821 | + | Transposase | |
| tniB | TnPfu1 | 1824-2732 | + | Accessory Gene | |
| tniQ | TnPfu1 | 2729-3946 | + | Accessory Gene | Target Site Selection |
| tniR | TnPfu1 | 4008-4631 | + | Accessory Gene | Resolvase |
| intI3 | TnPfu1 | 4733-5773 | - | Integron Integrase | Class 3 |
| AAC(6')-31 (ARO:3002585) | TnPfu1 | 5949-6467 | + | Passenger Gene | Antibiotic Resistance |
| FosX (ARO:3000198) | TnPfu1 | 6585-6941 | + | Passenger Gene | Antibiotic Resistance |
| bla IMP-1 (ARO:3002192) | TnPfu1 | 7006-7746 | + | Passenger Gene | Antibiotic Resistance |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniA | TniA | TnPfu1 | 559 | 142-1821 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | can be extended upstream by 12 amino acids| identical to tniA (Tn1721 and In2)| 25% amino acid sequence identity to TnsB from Tn7 | ||||||||||
| Protein Sequence: | MATDTPRIPE QGVATLPDEA WERARRRAEI ISPLAQSETV GHEAADMAAQ ALGLSRRQVY VLIRRARQGS GLVTDLVPGQ SGGGKGKGRL PEPVERVIHE LLQKRFLTKQ KRSLAAFHRE VTQVCKAQKL RVPARNTVAL RIASLDPRKV IRRREGQDAA RDLQGVGGEP PAVTAPLEQV QIDHTVIDLI VVDDRDRQPI GRPYLTLAID VFTRCVLGMV VTLEAPSAVS VGLCLVHVAC DKRPWLEGLN VEMDWQMSGK PLLLYLDNAA EFKSEALRRG CEQHGIRLDY RPLGQPHYGG IVERIIGTAM QMIHDELPGT TFSNPDQRGD YDSENKAALT LRELERWLTL AVGTYHGSVH NGLLQPPAAR WAEAVARVGV PAVVTRATSF LVDFLPILRR TLTRTGFVID HIHYYADALK PWIARRERWP SFLIRRDPRD ISRIWVLEPE GQHYLEIPYR TLSHPAVTLW EQRQALAKLR QQGREQVDES ALFRMIGQMR EIVTSAQKAT RKARRDADRR QHLKTSARPD KPVPPDTDIA DPQADNLPPA KPFDQIEEW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniB | TniB | TnPfu1 | 302 | 1824-2732 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Sequence Family: | ATP binding protein? | ||||||||||
| Comment: | similar function to Tn7 tnsC and MuB | ||||||||||
| Protein Sequence: | MDEYPIIDLS HLLPAAQGLA RLPADERVQR LRADRWIGYP RAVEALNRLE ALYAWPNKQR MPNLLLVGPT NNGKSMIVEK FRRTHPASAD ADQEHIPVLV VQMPSEPSVI RFYVALLAAM GAPLRPRPRL PEMEQLALAL LRKVGVRMLV IDELHNVLAG NSVNRREFLN LLRFLGNELR IPLVGVGTRD AYLAIRSDDQ LENRFEPMLL PVWEANDDCC SLLASFAASL PLRRPSSIAT LDMARYLLTR SEGTIGELAH LLMAAAVAAV ESGEEAINHR TLSMADYTGP SERRRQFERE LM |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniQ | TniQ | TnPfu1 | 405 | 2729-3946 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Target Site Selection | ||||||||||
| Comment: | similar function to Tn7 tnsC and MuB | ||||||||||
| Protein Sequence: | VKPAPRWPLH PAPKEGEALS SWLNRVALCY HMELPELLEH DLGHSQVDDL DTAPPLSLLA MLSQRSGIDP DRLRSMSFAG WVPWLLDSLD DQIPDALETY AFQLSVLLPK LHRKKRSITR WRAWLPSQPI HCACPLCLSD PDNQAVLLAW KLPLMLSCPL HGCWLESYWG VPGRFLGWEN ADAEPRTASD AIAAMDQRTW QALTTGHVEL PCRRIHAGLW FRLLRTLLDE LNTPLSACGT CAGYPRQVWE GCGHPLRAGQ SLWRPYETLN PVVRLQMLEA AATAISLIEV RDISPPGEQA KLFWSEPQTG FTSGLPTKAP KPEPINHWQR AVQAIDEAII EARHNPETAR SLFALASYGR RDPASLERLR ATFVKEGIPP EFLSHYLPDA PFACLKQNDG LSDKF |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniR | TniR | TnPfu1 | 207 | 4008-4631 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Comment: | also called tniR | ||||||||||
| Protein Sequence: | MLIGYMRVSK ADGSQSTDLQ RDALIVAGVS PAHLYEDMAS GRRDDRPGLA ACLKALREGD TLIVWKLDRL GRDLRHLINT VHDLTARSVG LKVLAGHGAA VDTTTAAGKL VFGIFAALAE FERELISERT VAGLVSARAR GRKGGRPFKM TATKLRLAMA SMGQPETKVG NLCEELGITR QTLYRHVSPK GELRPDGVKL LSRGSAA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | intI3 | IntI3 | TnPfu1 | 346 | 4733-5773 | - |
|---|
| Class: | Integron Integrase | ||||||||||
| Subclass: | Class 3 | ||||||||||
| Sequence Family: | Class 3 Integron Tyrosine Integrase | ||||||||||
| Transposase Chemistry: | Tyrosine | ||||||||||
| Protein Sequence: | MNRYNGSAKP DWVPPRSIKL LDQVRERVRY LHYSLQTEKA YVYWAKAFVL WTARSHGGFR HPREMGQAEV EGFLTMLATE KQVAPATHRQ ALNALLFLYR QVLGMELPWM QQIGRPPERK RIPVVLTVQE VQTLLSHMAG TEALLAALLY GSGLRLREAL GLRVKDVDFD RHAIIVRSGK GDKDRVVMLP RALVPRLRAQ LIQVRAVWGQ DRATGRGGVY LPHALERKYP RAGESWAWFW VFPSAKLSVD PQTGVERRHH LFEERLNRQL KKAVVQAGIA KHVSVHTLRH SFATHLLQAG TDIRTVQELL GHSDVSTTMI YTHVLKVAAG GTSSPLDALA LHLSPG |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | AAC(6')-31 (ARO:3002585) | AAC(6')-31 | TnPfu1 | 172 | 5949-6467 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | AAC(6') (ARO:3000345) | ||||||||||
| Transposase Chemistry: | aminoglycoside N(6')-acetyltransferase | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | 99% identity to reference sequence ARO:3002585 (bit score: 355) | ||||||||||
| Protein Sequence: | MTEHDLPMLH DWLNRPHIVE WWGGEETRPT LAEVLEQYLP SALAKESVTP YIAMLDEEPI GYAQSYIALG SGDGWWEDET DPGVRGIDQF LANPSQLGKG LGTKLVCALV EMLFKDAEVT KIQTDPSPNN LRAIRCYEKA GFVAQRTINT PDGPAVYMVQ TRQAFEQARS AV |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | FosX (ARO:3000198) | FosX | TnPfu1 | 118 | 6585-6941 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | fosfomycin thiol transferase (ARO:3000133) | ||||||||||
| Target: | fosfomycin (ARO:0000025) | ||||||||||
| Comment: | loose match to reference sequence for ARO:3000198 (53% identity, bitscore: 142) | ||||||||||
| Protein Sequence: | MTTFLCDGLG AREVYDSAGH NYSLSREKFF VLGGVWLAAM EGVPPSERSY QHVAFRVSES DLAVYQARLG SLGVEIRPPR PRVNGEGLSL YFYDFDNHLF ELHTGTLEQR LARYGAGR |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | bla IMP-1 (ARO:3002192) | Bla IMP-1 | TnPfu1 | 246 | 7006-7746 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | IMP beta-lactamase (ARO:3000020) | ||||||||||
| Target: | cephalosporin (ARO:0000032)||penem (ARO:3003706)||penam (ARO:3000008)||cephamycin (ARO:0000044)||carbapenem (ARO:0000020) | ||||||||||
| Comment: | 100% identical to reference sequence forARO:3002192 | ||||||||||
| Protein Sequence: | MSKLSVFFIF LFCSIATAAE SLPDLKIEKL DEGVYVHTSF EEVNGWGVVP KHGLVVLVNA EAYLIDTPFT AKDTEKLVTW FVERGYKIKG SISSHFHSDS TGGIEWLNSR SIPTYASELT NELLKKDGKV QATNSFSGVN YWLVKNKIEV FYPGPGHTPD NVVVWLPERK ILFGGCFIKP YGLGNLGDAN IEAWPKSAKL LKSKYGKAKL VVPSHSEVGD ASLLKLTLEQ AVKGLNESKK PSKPSN |
||||||||||
1. Yamamoto M, Matsumura Y, Gomi R, Matsuda T, Nakano S, Nagao M, Tanaka M, Ichiyama S.
Molecular Analysis of a bla(IMP-1)-Harboring Class 3 Integron in Multidrug-Resistant Pseudomonas fulva.. Antimicrob Agents Chemother. 2018 Jul 27;62(8):e00701-18. doi: 10.1128/AAC.00701-18. Print 2018 Aug. PubMed ID:
29784850
.