Mobile element type:
Transposon
Name:
TnMex22
Synonyms:
Tn7165
Accession:
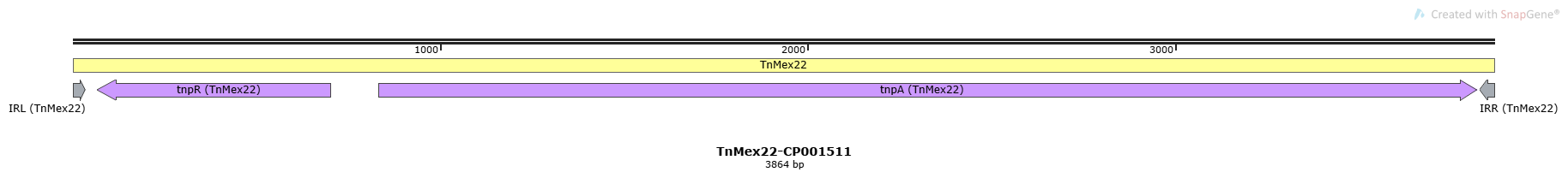
TnMex22-CP001511
Family:
Tn3
Group:
Tn163
First isolate:
Yes
Partial:
ND
Evidence of transposition:
Yes
Host Organism:
Methylobacterium extorquens AM1
Date of Isolation:
2009
Country:
England
Molecular Source:
megaplasmid
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-36 | + | 36 | GGCCCCTGAA CATTAAAGGG GCACGGATAT ACGGTA |
| IRR | 3829-3864 | - | 36 | GGCCCCTGAA CATTAAAGGG GCACGGATAT ACGGTA |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| tnpR | TnMex22 | 68-700 | - | Accessory Gene | Resolvase |
| tnpA | TnMex22 | 832-3822 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | TnMex22 | 210 | 68-700 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MTVLIGYARV SKADGSQVHD LQRDALRKAG VDPEHIYEDA ASGRSDDRPG LEACLKALRA GDTLAVWKLD RLGRDLRHLV NLVGDLTRRH VGLKVLSGEG ASIDTTTANG RLVFAIFAGL AEFERELISE RTKAGLAAAR ARGRKGGRPF KMTPAKLRLA QAAMGNPETC VADLCVELGI TRQTLYRFVG PRGELRPDGE KLLAQRRMRA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | TnMex22 | 996 | 832-3822 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MPRRLILTDA ERRTILALPT DEATLIRHWS LDDQDLALLD TRRRDDTRLG LALQLCALRY PGRLIRPGET IPEAAAVFLA DQLGGDPDAL ASFARRAPTR YEQLTILRRR FGFTDLCRPL RGDLVAFARG IALAVAKDRL VVTALAEEMR RRRIVIPGIT VLERLAAQAC TEAEDALLAD VAGRLTPDLV IRMEALLTVG PLAMGPRHAR QSGISWLREP PGSAGTAAMR GLVDRLEAVR HVGVPATVLG GVPAHRIRRM AQEGRRLTAQ NFAQMRPSRR HATLAAFLHD TQTALTDAAI GMFEILVGRA FRQAEADREA HLTASVVAAA EALDFFAGFG DALVAHKGVG LSLDAAITTV ATWEALARAT AAAQANRQAR HGDDTIAFLR RHHGRIRAFA APFLTRFTFE AARPGMALVT AVSQLGEAWK AGRRSPGQAW IDAALSLLDR RWSRHVRAPD GTIDRKMLEI FLVVELKNRI TAGEVWVAGS RTYRALEEKL IPPQTFAIIK AEARVPVAIP VDVEIYLAEK AAALEGKLQA AARRLKTGRG ETRIGAKGLR VPAVRTAETE AAVALARQVA ATMPPIRLTD LMADVDRMTG FSALFEHLQT GRPPADRRVF LAALIAEATN LGFGKMALAC PGLTRRQLQQ VAIWHFREDT FALALARLVE AQHAAPFSAT FGSHAIASSD GQHIYLGDGG EIAGGVNGHY GSDPITKLYT TISGRYAPFH VKIIAATASE AVHVLDALLE TEAGAAVTRH HVDGGGVSDL VFALCHGLGF AFVPRIPDLD GRCLYGFAPA RHYGVLQSVM GERLDAGLIR RHWDDILRLL TSLRTRTVSA SLVLRQLSAT PRQSGLVQAL RQMGRVERTL FTLDWIGDEQ LRKGTTAELN KGERRNGLVR AVNLHRLGRF RDRSQDSLAI RASALNLVVT AIIYWNTIYT GRVVDALRAR GALLPDHLLT GLSPLGWEHI GLTGDYLWEE TPGIDQTGFR AIPITP |
||||||||||
1. Vuilleumier S, Chistoserdova L, Lee MC, Bringel F, Lajus A, Zhou Y, Gourion B, Barbe V, Chang J, Cruveiller S, Dossat C, Gillett W, Gruffaz C, Haugen E, Hourcade E, Levy R, Mangenot S, Muller E, Nadalig T, Pagni M, Penny C, Peyraud R, Robinson DG, Roche D, Rouy Z, Saenampechek C, Salvignol G, Vallenet D, Wu Z, Marx CJ, Vorholt JA, Olson MV, Kaul R, Weissenbach J, Medigue C, Lidstrom ME.
Methylobacterium genome sequences: a reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources.. PLoS One. 2009;4(5):e5584. doi: 10.1371/journal.pone.0005584. Epub 2009 May 18. PubMed ID:
19440302
.