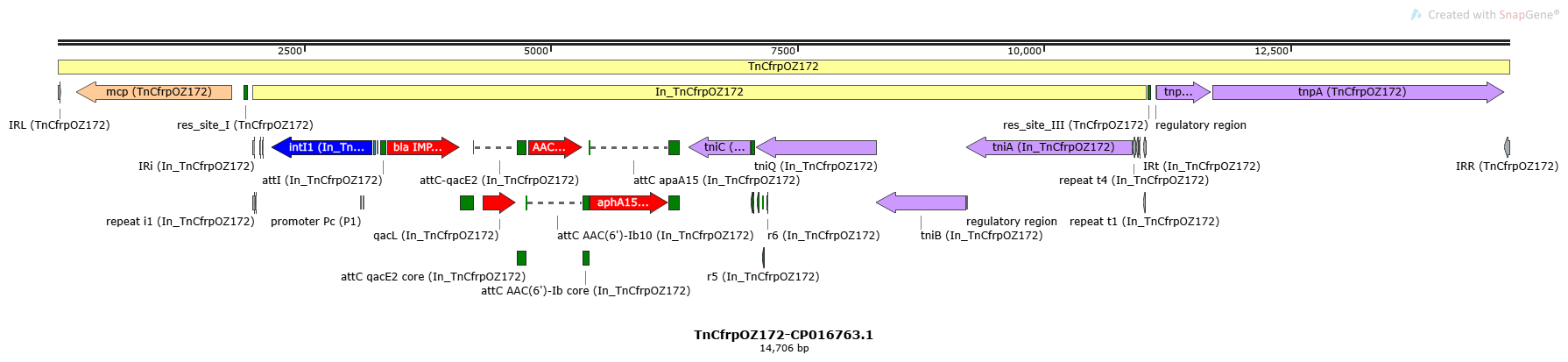
Mobile element type:
Transposon
Name:
TnCfrpOZ172
Synonyms:
Tn7154
Accession:
TnCfrpOZ172-CP016763.1
Family:
Tn3
Group:
Tn21
First isolate:
Yes
Partial:
ND
Evidence of transposition:
Yes
Host Organism:
Citrobacter freundii strain B38
Date of Isolation:
2001
Country:
China
Molecular Source:
plasmid pOZ172
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-38 | + | 38 | GGGGGAACCG CAGAATTCGG AAAAAATCGT ACGCTAAG |
| IRR | 14670-14706 | - | 37 | GGGGAGCCCG CAGAATTCGG AAAAAATCGT ACGCTAA |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res_site_I | TnCfrpOZ172 | 1886-1925 | + | 40 | GCGCATGTCA ATCTAGGCTA TACCCTAACT TGATGTCAGG |
| attI | In_TnCfrpOZ172 | 3269-3324 | + | 56 | TGATGTTATG GAGCAGCAAC GATGTTACGC AGCAGGGCAG TCGCCCTAAA ACAAAG |
| attC bla IMP-4 core | In_TnCfrpOZ172 | 4084-4216 | + | 133 | TTCTAACAAG TCGTTGCAGC ATCGTGCGCT GCGCGCACTG GACAGTTTTT AAGTCGCGGT TTTATGGTTT TGCTTCGCAA AAATATTCCA TAAAACCACA ACTTAAAAAC TGCCGCTGAA CTCGGCGTTA GAT |
| attC-qacE2 | In_TnCfrpOZ172 | 4211-4216 , 4655-4747 | + | 99 | TTAGATATCT AACCATTCCG TCGAGAGGGA CCGCCCACAA GCTGCGCTTG CGGGTTCCCT TCGCGGCTTC GCCGCTACGG CGGCCCCTCA CGTCAAACG |
| attC qacE2 core | In_TnCfrpOZ172 | 4655-4747 | + | 93 | ATCTAACCAT TCCGTCGAGA GGGACCGCCC ACAAGCTGCG CTTGCGGGTT CCCTTCGCGG CTTCGCCGCT ACGGCGGCCC CTCACGTCAA ACG |
| attC AAC(6')-Ib10 | In_TnCfrpOZ172 | 4747-4753 , 5321-5386 | + | 73 | GTTAGGCGCC TAACCCTTCC ATCGAGGGGG ACGTCCAAGG GCTGGCGCCC TTGGCCGCCC CTCATGTCAA ACG |
| attC AAC(6')-Ib core | In_TnCfrpOZ172 | 5321-5386 | + | 66 | GCCTAACCCT TCCATCGAGG GGGACGTCCA AGGGCTGGCG CCCTTGGCCG CCCCTCATGT CAAACG |
| attC apaA15 | In_TnCfrpOZ172 | 5387-5393 , 6197-6304 | + | 115 | TTAGACCGTC TAACAATTCG TTCAAGCCGA GATCGCTTCG CGGCCGCGGA GTTGTTCTGT AAATTGTCAC AACGCCGCGG CCGCAAAGCG CTCCGGCTTA ACTCAGGCGT TGAAC |
| attC aphA15 core | In_TnCfrpOZ172 | 6197-6304 | + | 108 | GTCTAACAAT TCGTTCAAGC CGAGATCGCT TCGCGGCCGC GGAGTTGTTC TGTAAATTGT CACAACGCCG CGGCCGCAAA GCGCTCCGGC TTAACTCAGG CGTTGAAC |
| res | In_TnCfrpOZ172 | 7031-7065 | + | 35 | ACCCGTCACG TATACGTTCG ATTATGTGAC AGTGT |
| r1 | In_TnCfrpOZ172 | 7033-7046 | - | 14 | CGTATACGTG ACGG |
| r2 | In_TnCfrpOZ172 | 7049-7062 | + | 14 | CGATTATGTG ACAG |
| r3 | In_TnCfrpOZ172 | 7093-7106 | - | 14 | GGGTTAAGTG ACAG |
| r4 | In_TnCfrpOZ172 | 7144-7157 | + | 14 | TCGTAATGTG ACAA |
| r5 | In_TnCfrpOZ172 | 7149-7162 | - | 14 | AGTTTTTGTC ACAT |
| r6 | In_TnCfrpOZ172 | 7194-7208 | + | 15 | GCGCAAACGT TCCAG |
| res_site_III | TnCfrpOZ172 | 11051-11081 | + | 31 | CGAAGGTTAT AGATTTCAGC CTGACAGAAA T |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| mcp | TnCfrpOZ172 | 190-1767 | - | Passenger Gene | Other |
| intI1 | In_TnCfrpOZ172 | 2175-3188 | - | IntegronIntegrase | Class 1 |
| bla IMP-4 (ARO:3002195) | In_TnCfrpOZ172 | 3341-4081 | + | Passenger Gene | Antibiotic Resistance |
| qacL (ARO:3005098) | In_TnCfrpOZ172 | 4313-4645 | + | Passenger Gene | Antibiotic Resistance |
| AAC(6')-Ib4 (ARO:3002577) | In_TnCfrpOZ172 | 4772-5326 | + | Passenger Gene | Antibiotic Resistance |
| aphA15 (ARO:3004675) | In_TnCfrpOZ172 | 5397-6191 | + | Passenger Gene | Antibiotic Resistance |
| tniC | In_TnCfrpOZ172 | 6401-7024 | - | Accessory Gene | Resolvase |
| tniQ | In_TnCfrpOZ172 | 7086-8303 | - | AccessoryGene | Target Site Selection |
| tniB | In_TnCfrpOZ172 | 8300-9208 | - | Accessory Gene | |
| tniA | In_TnCfrpOZ172 | 9211-10890 | - | Transposase | |
| tnpR | TnCfrpOZ172 | 11143-11703 | + | Accessory Gene | Resolvase |
| tnpA | TnCfrpOZ172 | 11707-14673 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | mcp | Mcp | TnCfrpOZ172 | 525 | 190-1767 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Comment: | methyl-accepting chemotaxis protein | ||||||||||
| Protein Sequence: | MRQNLPVTGR NLELPKDANI LSTTSPQSHI TYVNPDFIKI SGFTEEELLG QPHNIVRHPD MPPAAFEHMW STLKSGRSWM GLVKNRCKNG DHYWVSAYVT PIAKNGSIVE YQSVRTKPEP EQVLAAEKLY AQLRSGKAAR PKLAASFSVK ILLLIWGSII SSAMAAGMLT DTSISSLLLA TLMSGSLSSV SVLAILSPLG RLVERARNIS NNPLSQSLYT GRTDEFGQIE FALRMMQAET GAIVGRIGDA SNRLSEHTRG LLKDIESSNV LTVEQQAETD QIATAVNQMV ASIQEVASNA QHAADAAGRA DTETASGQRL VAHTSQSITA LEGEIRQATQ VIHELEGQSN EISKVLDVIR GIAEQTNLLA LNAAIEAARA GEQGRGFAVV ADEVRSLAAR TQQSTTDIQS MISALQERAQ SAVTVMEQSS RQAHTSVAHA EEAATALDGI GQRVNEITDM NAQIATAVEQ QGAVSEDINR SIINIRDAAD TNVQTGQNNL QSAKSVAQLT SALSELAKQF WEKRG |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | intI1 | IntI1 | In_TnCfrpOZ172 | 337 | 2175-3188 | - |
|---|
| Class: | IntegronIntegrase | ||||||||||
| Subclass: | Class 1 | ||||||||||
| Sequence Family: | Class 1 Integron Tyrosine Integrase | ||||||||||
| Transposase Chemistry: | Tyrosine | ||||||||||
| Protein Sequence: | MKTATAPLPP LRSVKVLDQL RERIRYLHYS LPTEQAYVNW VRAFIRFHGV RHPATLGSSE VEAFLSWLAN ERKVSVSTHR QALAALLFFY GKVLCTDLPW LQEIGRPRPS RRLPVVLTPD EVVRILGFLE GEHRLFAQLL YGTGMRISEG LQLRVKDLDF DHGTIIVREG KGSKDRALML PESLAPSLRE QLSRARAWWL KDQAEGRSGV ALPDALERKY PRAGHSWPWF WVFAQHTHST DPRSGVVRRH HMYDQTFQRA FKRAVEQAGI TKPATPHTLR HSFATALLRS GYDIRTVQDL LGHSDVSTTM IYTHVLKVGG AGVRSPLDAL PPLTSER |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | bla IMP-4 (ARO:3002195) | Bla IMP-4 | In_TnCfrpOZ172 | 246 | 3341-4081 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | IMP beta-lactamase (ARO:3000020) | ||||||||||
| Target: | carbapenem (ARO:0000020)||cephalosporin (ARO:0000032)||cephamycin (ARO:0000044)||penam (ARO:3000008)||penem (ARO:3003706) | ||||||||||
| Comment: | strict match to reference sequence for (ARO:3002195) (bitscore: 497)||Synonyms: | ||||||||||
| Protein Sequence: | MSKLSVFFIF LFCSIATAAE PLPDLKIEKL DEGVYVHTSF EEVNGWGVVP KHGLVVLVDA EAYLIDTPFT AKDTEKLVTW FVERGYKIKG SISSHFHSDS TGGIEWLNSQ SIPTYASELT NELLKKDGKV QAKNSFGGVN YWLVKNKIEV FYPGPGHTPD NLVVWLPERK ILFGGCFIKP YGLGNLGDAN LEAWPKSAKL LISKYGKAKL VVPSHSEAGD ASLLKLTLEQ AVKGLNESKK PSKLSN |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | qacL (ARO:3005098) | QacL | In_TnCfrpOZ172 | 110 | 4313-4645 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic efflux (ARO:0010000) | ||||||||||
| Sequence Family: | small multidrug resistance (SMR) antibiotic efflux pump (ARO:0010003) | ||||||||||
| Target: | disinfectingagents and antiseptics (ARO:3005386) | ||||||||||
| Comment: | subunit of the qac multidrug efflux pump||loose match to reference sequence for ARO:3005098 (bitscore:172) | ||||||||||
| Protein Sequence: | MKNWLFLATS IIFEVIATSA LKSSEGFTRL VPSFIVVAGY AAAFYFLSLT LKSIPVGIAY AVWSGLGIVL VTAIAWVLHG QKLDMWGFVG VGFIISGVAV LNLLSKASVH |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | AAC(6')-Ib4 (ARO:3002577) | AAC(6')-Ib4 | In_TnCfrpOZ172 | 184 | 4772-5326 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | AAC(6') (ARO:3000345) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | strict match to reference sequence for ARO:3002577 (bitscore: 380) | ||||||||||
| Protein Sequence: | MTNSNDSVTL RLMTEHDLAM LYEWLNRSHI VEWWGGEEAR PTLADVQEQY LPSVLAQESV TPYIAMLNGE PIGYAQSYVA LGSGDGWWEE ETDPGVRGID QSLANASQLG KGLGTKLVRA LVELLFNDPE VTKIQTDPSP SNLRAIRCYE KAGFERQGTV TTPDGPAVYM VQTRQAFERT RSDA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | aphA15 (ARO:3004675) | AphA15 | In_TnCfrpOZ172 | 264 | 5397-6191 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | APH(3') (ARO:3000126) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | strict match to reference sequence for ARO:3004675 (bitscore: 543) | ||||||||||
| Protein Sequence: | MTVALDEVSE LKNLLSPLLD ECTFEEVEYG QSDARVIRVL FPDRNTAYLK YASGSSAQEI LQEHQRTRWL RTRALVPEVI SYVSTSTVTI LLTKALIGHN AADAADADPV IVVAEMARAL RDLHSISPDD CPFDERLHLR LKLASGRLEA GLVDEEDFDH ARQGMLARDV YEQLFIQMPG AEQLVVTHGD ACPENFIFQG NAFVGFIDCG RVGLADKYQD LALASRNIDA VFGPELTNQF FIEYGEPNPN IAKIEYYRIL DEFF |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniC | TniC | In_TnCfrpOZ172 | 207 | 6401-7024 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Comment: | identical to tniR (Tn1721) | ||||||||||
| Protein Sequence: | MLIGYMRVSK ADGSQSTNLQ RDALIAAGVS LAHLYEDLAS GRRDDRPGLA ACLKALREGD TLIVWKLDRL GRDLRHLINT VHDLTARSVG LKVLTGHGAA VDTTTAAGKL VFGIFAALAE FERELISERT VAGLISARAR GRKGGRPFKM TAAKLRLAMA SMGQPETKVG DLCEELGITR QTLYRHVSPK GELRPDGVKL LSLGSAA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniQ | TniQ | In_TnCfrpOZ172 | 405 | 7086-8303 | - |
|---|
| Class: | AccessoryGene | ||||||||||
| Subclass: | Target Site Selection | ||||||||||
| Function: | is needed for transposition of Tn5053 | ||||||||||
| Protein Sequence: | MKSAPRWPLH PAPKEGEALS SWLNRVAACY QMDVHELLAH DLGHSQLDDL DTAPSLSLLT ALCQRSGVEL ERLRSMSLAG WVPWLLDSLD DSVPAALETY TFQCAVLLPK RTRKVRSITR WRAWLPSQTI RRACPQCLND PTNQAVLLVW QLPLMLSCPQ HGCWLESYWG MPGRYLQWEI ADAAPRPADD AIACMDRRTW QALTTGFVEL PRRRVHAGLW FRLLRTLLDE LNTPLSHCGS CSASIRHVWE RCGHPLRAGQ SLWRPYEILA PAVQWQMLEA AATAITLIES KALIPRGEQA ALFQTEPHTG FTNGLPAKVP KPEPINHWQR AAQAIDEAII EARHNPETAR LLFTLASYGR RDPESLERLR ATFTKEGIPP EFLSHYEPEW PFAGLRLNDG LSDSF |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniB | TniB | In_TnCfrpOZ172 | 302 | 8300-9208 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Function: | is needed for transposition of Tn5053 | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Comment: | homologous to TnsC protein of Tn7 putative ATP-binding protein | ||||||||||
| Protein Sequence: | MDEYPVIDLS HLLPAAQGLA RLPADERIQR IRADRWIGYP RAVEALNRLE TLYAWPNKQR MPNLLLVGPT NNGKSMIVEK FRRAHPVGTD ADQEHIPVLV VQMPSEPSVI RFYVALLAAM GAPLRPRPRL PEMEQLTLAL LRKVGVRMLV IDELHNVLAG NSVNRREFLN LLRFLGNELR IPLVGVGTRE AYLAIRSDDQ LENRFEPMLL PPWEANEDCC SLLASFAASL PLRRPSPIAT LDMARYLLTR SEGTIGELAH LLVAAAVAAV ESGEEAINHR TLSMADYIGP SERRRQFERE LM |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniA | TniA | In_TnCfrpOZ172 | 559 | 9211-10890 | - |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | homologous to TnsB of Tn7 | ||||||||||
| Protein Sequence: | MTSDTPPIAA QGVATLPDEA WAQARHRTEI IGPLAALEVV GHEAADEAAQ ALGLSRRQVY VLIRRARQGT GLVTDLTPGR SGGGKGKGRL PEPVERIIRE LLQKRFLTKQ KRSLAAFHRE VAQACKTQKL PVPARNTVAQ RIAGLHPAKI ARSRGGQDAA RPLQGAGGIP PEVTMPLEQV QIDHTVIDLI VVDERDRQPI GRPYLTLAID VFTRCVLGMV VTLEAPSAVS VGLCLAHAAC DKRPWLEGLN VEMDWPMSGK PRLLYLDNAA EFKSEALRRG CEQHGIRLDY RPPGQPHYGG IVERIIGTAM QMIHDELPGT TFSNPGQRGE YDSEKMATLT LRELERWLAL AVGTYHGSVH NGLLQPPAAR WAEAVERVGV PAVVTRPTAF LVDFLPVIRR TLTRTGFVID HIHYYADALK PWIARRERLP AFLIRRDPRD ISRIWVLEPE GQHYLEIHYR TLSHPAVTLW EQRQALAKLR QLGREQVDES ALFRMIGQMR EIVTTAQKAT RKARRDADRR QHLKTSEPPA KPIPPDVDMA DPQADNLPPA KPFDQIEEW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | TnCfrpOZ172 | 186 | 11143-11703 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MQGHRIGYVR VSSFDQNPER QLEQTQVSKV FTDKASGKDT QRPQLEALLS FVREGDTVVV HSMDRLARNL DDLRRLVQKL TQRGVRIEFL KEGLVFTGED SPMANLMLSV MGAFAEFERA LIRERQREGI ALAKQRGAYR GRKKALSDEQ AATLRQRATA GEPKAQLARE FNISRETLYQ YLRTDD |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | TnCfrpOZ172 | 988 | 11707-14673 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MPRRLILSAT ERDTLLALPE SQDDLIRYYT FNDSDLSLIR QRRGDANRLG FAVQLCLLRY PGYALGTDSE LPEPVILWVA KQVQAEPASW AKYGERDVTR REHAQELRTY LQLAPFGLSD FRALVRELTE LAQQTDKGLL LAGQALESLR QKRRILPALS VIDRACSEAI ARANRRVYRA LVEPLTDSHR AKLDELLKLK AGSSITWLTW LRQAPLKPNS RHMLEHIERL KTFQLVDLPE GLGRHIHQNR LLKLAREGGQ MTPKDLGKFE PQRRYATLAA VVLESTATVI DELVDLHDRI LVKLFSGAKH KHQQQFQKQG KAINDKVRLY SRIGQALLEA KESGSDPYAA IEAVIPWDEF TESVSEAELL ARPEGFDHLH LVGENFATLR RYTPALLEVL ELRAAPAAQG VLAAVQTLRE MNADNLRKVP ADAPTAFIKP RWKPLVITPE GLDRKFYEIC ALSELKNALR SGDIWVKGSR QFRDFDDYLL PAEKFAALKR EQALPLAINP NSDQYLEERL QLLDEQLATV TRLAKDNELP DAILTESGLK ITPLDAAVPD RAQALIDQTS QLLPRIKITE LLMDVDDWTG FSRHFTHLKD GAEAKDRTLL LSAILGDAIN LGLTKMAESS PGLTYAKLSW LQAWHIRDET YSAALAELVN HQYRHAFAAH WGDGTTSSSD GQRFRAGGRG ESTGHVNPKY GSEPGRLFYT HISDQYAPFS TRVVNVGVRD STYVLDGLLY HESDLRIEEH YTDTAGFTDH VFALMHLLGF RFAPRIRDLG ETKLYVPQGV QAYPTLRPLI GGTLNIKHVR AHWDDILRLA SSIKQGTVTA SLMLRKLGSY PRQNGLAVAL RELGRIERTL FILDWLQSVE LRRRVHAGLN KGEARNSLAR AVFFNRLGEI RDRSFEQQRY RASGLNLVTA AIVLWNTVYL ERATQGLVEA GKPVDGELLQ FLSPLGWEHI NLTGDYVWRQ SRRLEDGKFR PLRMPGKP |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| In_TnCfrpOZ172-CP016763.1 | In_TnCfrpOZ172 | integron | 1973-11033 | + | 9061 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRi | In_TnCfrpOZ172 | 1973-2005 | + | 33 | TGTCGTTTTC AGAAGACGGC TGCACTGAAC GTC |
| repeat i1 | In_TnCfrpOZ172 | 1981-1999 | + | 19 | TCAGAAGACG GCTGCACTG |
| repeat i2 | In_TnCfrpOZ172 | 2004-2022 | + | 19 | TCAGAAGCCG ACTGCACTA |
| repeat i3 | In_TnCfrpOZ172 | 2046-2064 | + | 19 | TCAGGCAACG ACGGGCTGC |
| repeat i4 | In_TnCfrpOZ172 | 2074-2092 | + | 19 | TCAGCGGACG CAGGGAGGA |
| repeat t4 | In_TnCfrpOZ172 | 10906-10924 | - | 19 | TCAATACTCG TGTGCACCA |
| repeat t3 | In_TnCfrpOZ172 | 10938-10956 | - | 19 | TCAGGAGTCG TCTGCACCA |
| repeat t2 | In_TnCfrpOZ172 | 10967-10985 | + | 19 | TCAGGAGCCG TATGCACAC |
| repeat t1 | In_TnCfrpOZ172 | 11007-11025 | - | 19 | TCAGAAGACG ACCGCACCA |
| IRt | In_TnCfrpOZ172 | 11009-11033 | - | 25 | TGTCGTTTTC AGAAGACGAC CGCAC |
1. Hawkey PM, Xiong J, Ye H, Li H, M'Zali FH.
Occurrence of a new metallo-beta-lactamase IMP-4 carried on a conjugative plasmid in Citrobacter youngae from the People's Republic of China.. FEMS Microbiol Lett. 2001 Jan 1;194(1):53-7. doi: 10.1111/j.1574-6968.2001.tb09445.x. PubMed ID:
11150665
.
2. Xiong J, Deraspe M, Iqbal N, Ma J, Jamieson FB, Wasserscheid J, Dewar K, Hawkey PM, Roy PH. Genome and Plasmid Analysis of blaIMP-4-Carrying Citrobacter freundii B38.. Antimicrob Agents Chemother. 2016 Oct 21;60(11):6719-6725. doi: 10.1128/AAC.00588-16. Print 2016 Nov. PubMed ID: 27572407 .
2. Xiong J, Deraspe M, Iqbal N, Ma J, Jamieson FB, Wasserscheid J, Dewar K, Hawkey PM, Roy PH. Genome and Plasmid Analysis of blaIMP-4-Carrying Citrobacter freundii B38.. Antimicrob Agents Chemother. 2016 Oct 21;60(11):6719-6725. doi: 10.1128/AAC.00588-16. Print 2016 Nov. PubMed ID: 27572407 .