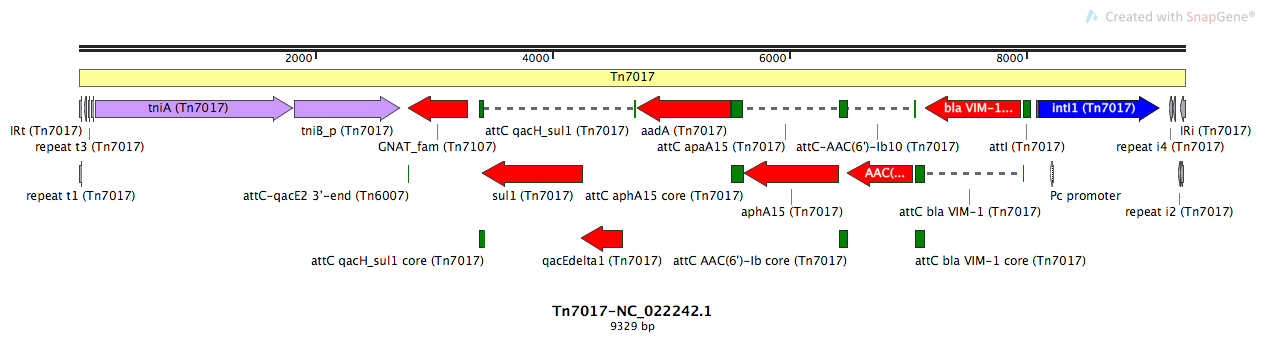
Mobile element type:
Transposon
Name:
Tn7017
Synonyms:
Accession:
Tn7017-NC_022242.1
Family:
Tn402
Group:
First isolate:
Yes
Partial:
ND
Evidence of transposition:
ND
Host Organism:
Achromobacter xylosoxidans subsp. denitrificans
Date of Isolation:
2014
Country:
Italy
Molecular Source:
plasmid pAX22
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRt | 1-33 | + | 33 | TGTCATTTTC AGAAGACGAC TGCACCAGTT GAT |
| repeat t1 | 9-27 | + | 19 | TCAGAAGACG ACTGCACCA |
| repeat t2 | 49-67 | - | 19 | TCAGGAGCTG GCTGCACAA |
| repeat t3 | 78-97 | + | 20 | TCAGAAGTGA TCTGCACCAA |
| repeat t4 | 110-128 | + | 19 | TCAATACTCG TGTGCACCA |
| repeat i4 | 9210-9228 | - | 19 | TCAGCGGACG CAGGGAGGA |
| repeat i3 | 9238-9256 | - | 19 | TCAGGCAACG ACGGGCTGC |
| repeat i2 | 9280-9298 | - | 19 | TCAGAAGCCG ACTGCACTA |
| IRi | 9297-9329 | - | 33 | TGTCGTTTTC AGAAGACGGC TGCACTGAAC GTC |
| repeat i1 | 9303-9321 | - | 19 | TCAGAAGACG GCTGCACTG |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| attC qacH_sul1 | Tn7017 | 3380-3413 , 4691-4697 | + | 41 | CCGCTAGCGG GCGGCCGGAA GGTGAATGCT AGGCATCTAA C |
| attC qacH_sul1 core | Tn7017 | 3380-3413 | + | 34 | CCGCTAGCGG GCGGCCGGAA GGTGAATGCT AGGC |
| attC qacH_sul1 | Tn7017 | 5507-5614 , 6418-6424 | + | 115 | GTTTAACGCC TGAGTTAAGC CGGAGCGCTT TGCGGCCGCG GCGTTGTGAC AATTTACAGA ACAACTCCGC GGCCGCGAAG CGATCTCGGC TTGAACGAAT TGTTAGACGG TCTAA |
| attC aphA15 core | Tn7017 | 5507-5614 | + | 108 | GTTTAACGCC TGAGTTAAGC CGGAGCGCTT TGCGGCCGCG GCGTTGTGAC AATTTACAGA ACAACTCCGC GGCCGCGAAG CGATCTCGGC TTGAACGAAT TGTTAGAC |
| attC-AAC(6')-Ib10 | Tn7017 | 6425-6490 , 7058-7064 | + | 73 | CGTTTGACAT GAGGGGCGGC CAAGGGCGCC AGCCCTTGGA CGTCCCCCTC GATGGAAGGG TTAGGCGCCT AAC |
| attC AAC(6')-Ib core | Tn7017 | 6425-6490 | + | 66 | CGTTTGACAT GAGGGGCGGC CAAGGGCGCC AGCCCTTGGA CGTCCCCCTC GATGGAAGGG TTAGGC |
| attC qacH_sul1 | Tn7017 | 7064-7138 , 7972-7978 | + | 82 | CGCCTGAGTT GAGGGGCTTT TGCGGAGTAG GCGCAGCCTG CGTAGCAAAA GTCCCGCTCC AACGATTTGT TATGCGCATA AC |
| attC bla VIM-1 core | Tn7017 | 7064-7138 | + | 75 | CGCCTGAGTT GAGGGGCTTT TGCGGAGTAG GCGCAGCCTG CGTAGCAAAA GTCCCGCTCC AACGATTTGT TATGC |
| attI | Tn7017 | 7978-8033 | + | 56 | CTTTGTTTTA GGGCGACTGC CCTGCTGCGT AACATCGTTG CTGCTCCATA ACATCA |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| tniA | Tn7017 | 142-1821 | + | Transposase | |
| tniB_p | Tn7017 | 1824-2723 | + | Accessory Gene | |
| sul1 (ARO:3000410) | Tn7017 | 3408-4247 | - | Passenger Gene | Antibiotic Resistance |
| qacEdelta1 (ARO:3005010) | Tn7017 | 4241-4588 | - | Passenger Gene | Antibiotic Resistance |
| aadA (ARO:3002601) | Tn7017 | 4712-5503 | - | Passenger Gene | Antibiotic Resistance |
| aphA15 (ARO:3004675) | Tn7017 | 5620-6414 | - | Passenger Gene | Antibiotic Resistance |
| AAC(6')-Ib10 (ARO:3002581) | Tn7017 | 6485-7039 | - | Passenger Gene | Antibiotic Resistance |
| bla VIM-1 (ARO:3002271) | Tn7017 | 7147-7947 | - | Passenger Gene | Antibiotic Resistance |
| intI1 | Tn7017 | 8114-9127 | + | Integron Integrase | Class 1 |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniA | TniA | Tn7017 | 559 | 142-1821 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | can be extended upstream by 12 amino acids| identical to tniA (Tn1721 and In2)| 25% amino acid sequence identity to TnsB from Tn7 | ||||||||||
| Protein Sequence: | MATDTPRIPE QGVATLPDEA WERARRRAEI ISPLAQSETV GHEAADMAAQ ALGLSRRQVY VLIRRARQGS GLVTDLVPGQ SGGGKGKGRL PEPVERVIHE LLQKRFLTKQ KRSLAAFHRE VTQVCKAQKL RVPARNTVAL RIASLDPRKV IRRREGQDAA RDLQGVGGEP PAVTAPLEQV QIDHTVIDLI VVDDRDRQPI GRPYLTLAID VFTRCVLGMV VTLEAPSAVS VGLCLVHVAC DKRPWLEGLN VEMDWQMSGK PLLLYLDNAA EFKSEALRRG CEQHGIRLDY RPLGQPHYGG IVERIIGTAM QMIHDELPGT TFSNPDQRGD YDSENKAALT LRELERWLTL AVGTYHGSVH NGLLQPPAAR WAEAVARVGV PAVVTRATSF LVDFLPILRR TLTRTGFVID HIHYYADALK PWIARRERWP SFLIRRDPRD ISRIWVLEPE GQHYLEIPYR TLSHPAVTLW EQRQALAKLR QQGREQVDES ALFRMIGQMR EIVTSAQKAT RKARRDADRR QHLKTSARPD KPVPPDTDIA DPQADNLPPA KPFDQIEEW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniB_p | TniB_p | Tn7017 | 299 | 1824-2723 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Sequence Family: | ATP binding protein | ||||||||||
| Comment: | 3' deletion| similar function to Tn7 tnsC and MuB | ||||||||||
| Protein Sequence: | VDEYPIIDLS HLLPAAQGLA RLPADERIQR LRADRWIGYP RAVEALNRLE ALYAWPNKQR MPNLLLVGPT NNGKSMIVEK FRRTHPASSD ADQEHIPVLV VQMPSEPSVI RFYVALLAAM GAPLRPRPRL PEMEQLALAL LRKVGVRMLV IDELHNVLAG NSVNRREFLN LLRFLGNELR IPLVGVGTRD AYLAIRSDDQ LENRFEPMML PVWEANDDCC SLLASFAASL PLRRPSPIAT LDMARYLLTR SEGTIGELAH LLMAAAIVAV ESGEEAINHR TLSISTTSRA TKTSRSDCK |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | sul1 (ARO:3000410) | Sul1 | Tn7017 | 279 | 3408-4247 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic target replacement (ARO:0001002) | ||||||||||
| Sequence Family: | sulfonamide resistant sul (ARO:3004238) | ||||||||||
| Transposase Chemistry: | dihydropteroate synthase | ||||||||||
| Target: | sulfonamide antibiotic (ARO:3000282)||sulfone antibiotic (ARO:3003401) | ||||||||||
| Comment: | perfect match to reference sequence for ARO:3000410 | ||||||||||
| Protein Sequence: | MVTVFGILNL TEDSFFDESR RLDPAGAVTA AIEMLRVGSD VVDVGPAASH PDARPVSPAD EIRRIAPLLD ALSDQMHRVS IDSFQPETQR YALKRGVGYL NDIQGFPDPA LYPDIAEADC RLVVMHSAQR DGIATRTGHL RPEDALDEIV RFFEARVSAL RRSGVAADRL ILDPGMGFFL SPAPETSLHV LSNLQKLKSA LGLPLLVSVS RKSFLGATVG LPVKDLGPAS LAAELHAIGN GADYVRTHAP GDLRSAITFS ETLAKFRSRD ARDRGLDHA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | qacEdelta1 (ARO:3005010) | QacEdelta1 | Tn7017 | 115 | 4241-4588 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic efflux (ARO:0010000) | ||||||||||
| Sequence Family: | major facilitator superfamily (MFS) antibiotic efflux pump (ARO:0010002) | ||||||||||
| Target: | acridine dye (ARO:3000054)||quaternary ammonium salts | ||||||||||
| Comment: | subunit of the qac multidrug efflux pump||perfect match to reference sequence for ARO:3005010 (bitscore:219) | ||||||||||
| Protein Sequence: | MKGWLFLVIA IVGEVIATSA LKSSEGFTKL APSAVVIIGY GIAFYFLSLV LKSIPVGVAY AVWSGLGVVI ITAIAWLLHG QKLDAWGFVG MGLIIAAFLL ARSPSWKSLR RPTPW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | aadA (ARO:3002601) | AadA | Tn7017 | 263 | 4712-5503 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | ANT(3'')(ARO:3004275) | ||||||||||
| Transposase Chemistry: | aminoglycoside nucleotidyltransferase | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | perfect match to reference sequence for ARO:3002601||Synonyms: aadA1-pm, aadA, aadA1, aad(3'')(9) | ||||||||||
| Protein Sequence: | MREAVIAEVS TQLSEVVGVI ERHLEPTLLA VHLYGSAVDG GLKPHSDIDL LVTVTVRLDE TTRRALINDL LETSASPGES EILRAVEVTI VVHDDIIPWR YPAKRELQFG EWQRNDILAG IFEPATIDID LAILLTKARE HSVALVGPAA EELFDPVPEQ DLFEALNETL TLWNSPPDWA GDERNVVLTL SRIWYSAVTG RIAPKDVAAD WAMERLPAQY QPVILEARQA YLGQEEDRLA SRADQLEEFV HYVKGEITKV VGK |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | aphA15 (ARO:3004675) | AphA15 | Tn7017 | 264 | 5620-6414 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | APH(3') (ARO:3000126) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | strict match to reference sequence for ARO:3004675 (bitscore: 543) | ||||||||||
| Protein Sequence: | MTVALDEVSE LKNLLSPLLD ECTFEEVEYG QSDARVIRVL FPDRNTAYLK YASGSSAQEI LQEHQRTRWL RTRALVPEVI SYVSTSTVTI LLTKALIGHN AADAADADPV IVVAEMARAL RDLHSISPDD CPFDERLHLR LKLASGRLEA GLVDEEDFDH ARQGMLARDV YEQLFIQMPG AEQLVVTHGD ACPENFIFQG NAFVGFIDCG RVGLADKYQD LALASRNIDA VFGPELTNQF FIEYGEPNPN IAKIEYYRIL DEFF |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | AAC(6')-Ib10 (ARO:3002581) | AAC(6')-Ib10 | Tn7017 | 184 | 6485-7039 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | AAC(6') (ARO:3000345) | ||||||||||
| Transposase Chemistry: | aminoglycoside acetyltransferase | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | strict match to reference sequence for ARO:3002581 (bitscore: 377) | ||||||||||
| Protein Sequence: | MTNSNDSVTL RLMTEHDLAM LYEWLNRSHI VEWWGGEEAR PTLADVQEQY LPSVLAQESV TPYIAMLNGE PIGYAQSYVA LGSGDGWWEE ETDPGVRGID QSLANASQLG KGLGTKLVRA LVELLFNDPE VTKIQTDPSP SNLRAIRCYE KAGFERQGTV TTPDGPAVYM VQTRQAFERT RSDA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | bla VIM-1 (ARO:3002271) | Bla VIM-1 | Tn7017 | 266 | 7147-7947 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | VIM beta-lactamase (ARO:3000021) | ||||||||||
| Target: | carbapenem (ARO:0000020)||cephamycin (ARO:0000044)||penam (ARO:3000008)||penem (ARO:3003706)||cephalosporin (ARO:0000032) | ||||||||||
| Comment: | strict match to reference sequence for ARO:3002271 (bitscore: 536) | ||||||||||
| Protein Sequence: | MLKVISSLLV YMTASVMAVA SPLAHSGEPS GEYPTVNEIP VGEVRLYQIA DGVWSHIATQ SFDGAVYPSN GLIVRDGDEL LLIDTAWGAK NTAALLAEIE KQIGLPVTRA VSTHFHDDRV GGVDVLRAAG VATYASPSTR RLAEAEGNEI PTHSLEGLSS SGDAVRFGPV ELFYPGAAHS TDNLVVYVPS ANVLYGGCAV HELSSTSAGN VADADLAEWP TSVERIQKHY PEAEVVIPGH GLPGGLDLLQ HTANVVKAHK NRSVAE |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | intI1 | IntI1 | Tn7017 | 337 | 8114-9127 | + |
|---|
| Class: | Integron Integrase | ||||||||||
| Subclass: | Class 1 | ||||||||||
| Sequence Family: | Class 1 Integron Tyrosine Integrase | ||||||||||
| Transposase Chemistry: | Tyrosine | ||||||||||
| Protein Sequence: | MKTATAPLPP LRSVKVLDQL RERIRYLHYS LRTEQAYVNW VRAFIRFHGV RHPATLGSSE VEAFLSWLAN ERKVSVSTHR QALAALLFFY GKVLCTDLPW LQEIGRPRPS RRLPVVLTPD EVVRILGFLE GEHRLFAQLL YGTGMRISEG LQLRVKDLDF DHGTIIVREG KGSKDRALML PESLAPSLRE QLSRARAWWL KDQAEGRSGV ALPDALERKY PRAGHSWPWF WVFAQHTHST DPRSGVVRRH HMYDQTFQRA FKRAVEQAGI TKPATPHTLR HSFATALLRS GYDIRTVQDL LGHSDVSTTM IYTHVLKVGG AGVRSPLDAL PPLTSER |
||||||||||
1. Di Pilato V, Pollini S, Rossolini GM.
Characterization of plasmid pAX22, encoding VIM-1 metallo-beta-lactamase, reveals a new putative mechanism of In70 integron mobilization.. J Antimicrob Chemother. 2014 Jan;69(1):67-71. doi: 10.1093/jac/dkt311. Epub 2013 Aug 8. PubMed ID:
23928025
.