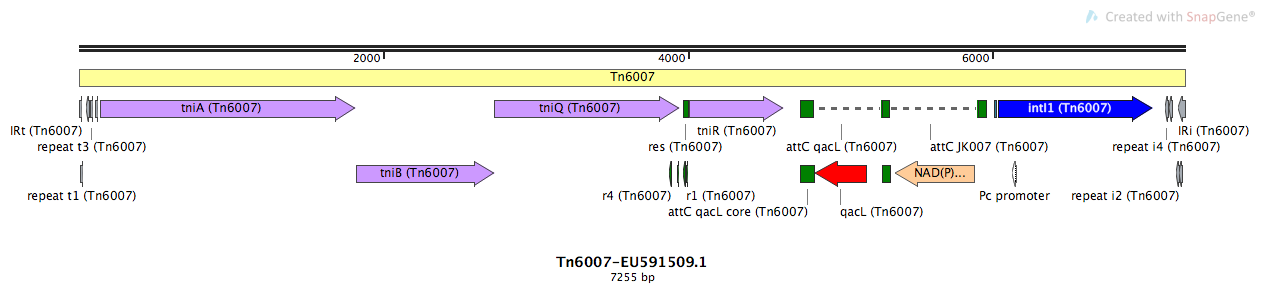
Mobile element type:
Transposon
Name:
Tn6007
Synonyms:
Accession:
Tn6007-EU591509.1
Family:
Tn402
Group:
First isolate:
ND
Partial:
ND
Evidence of transposition:
ND
Host Organism:
Enterobacter cloacaeJKB7
Date of Isolation:
None
Country:
None
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRt | 1-25 | + | 25 | TGTCATTTTC AGAAGACGAC TGCAC |
| repeat t1 | 9-27 | + | 19 | TCAGAAGACG ACTGCACCA |
| repeat t2 | 50-67 | - | 18 | TCAGGAGCCG ACTGCACA |
| repeat t4 | 110-128 | + | 19 | TCAATACTCG TGTGCACCA |
| repeat i4 | 7136-7154 | - | 19 | TCAGCGGACG CAGGGAGGA |
| repeat i3 | 7164-7182 | - | 19 | TCAGGCAACG ACGGGCTGC |
| repeat i2 | 7206-7224 | - | 19 | TCAGAAGCCG ACTGCACTA |
| IRi | 7223-7255 | - | 33 | TGTCGTTTTC AGAAGACGGC TGCACTGAAC GTC |
| repeat i1 | 7229-7247 | - | 19 | TCAGAAGACG GCTGCACTG |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| repeat t3 | Tn6007 | 78-97 | + | 20 | TCAGAAGTCA TCTGCACCAA |
| r4 | Tn6007 | 3877-3890 | - | 14 | TCGTAATGTG ACAA |
| r3 | Tn6007 | 3928-3941 | + | 14 | GGGTTAAGTG ACAA |
| res | Tn6007 | 3969-4003 | + | 35 | ACACTGTCAC ATAATCGAAC GTATACGTGA CGGGT |
| r2 | Tn6007 | 3972-3985 | - | 14 | CGATTATGTG ACAG |
| r1 | Tn6007 | 3988-4001 | + | 14 | CGTATACGTG ACGG |
| attC qacL (Tn6007) | Tn6007 | 4736-4828 , 5268-5273 | + | 99 | CGTTTGACGT GAGGGGCCGC CGTAGCGGCG AAGCCGCGAA GGGAACCCGC AAGCGCAGCT TGTGGGCGGT CCCTCTCGAC GGAATGGTTA GATATCTAA |
| attC qacL core | Tn6007 | 4736-4828 | + | 93 | CGTTTGACGT GAGGGGCCGC CGTAGCGGCG AAGCCGCGAA GGGAACCCGC AAGCGCAGCT TGTGGGCGGT CCCTCTCGAC GGAATGGTTA GAT |
| attC JK007 | Tn6007 | 5274-5327 , 5898-5903 | + | 60 | CGCTGGAGTT AAGCCGCGGC GCGTAGCGCC GTCGGCTTGA ACGACTTGTT ATACGTATAA |
| attC JK007 core | Tn6007 | 5274-5327 | + | 54 | CGCTGGAGTT AAGCCGCGGC GCGTAGCGCC GTCGGCTTGA ACGACTTGTT ATAC |
| attI | Tn6007 | 5904-5959 | + | 56 | CTTTGTTTTA GGGCGACTGC CCTGCTGCGT AACATCGTTG CTGCTCCATA ACATCA |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| tniA | Tn6007 | 144-1823 | + | Transposase | |
| tniB | Tn6007 | 1826-2734 | + | Accessory Gene | |
| tniQ | Tn6007 | 2731-3948 | + | Accessory Gene | Target Site Selection |
| tniR | Tn6007 | 4010-4633 | + | Accessory Gene | Resolvase |
| qacL (ARO:3005098) | Tn6007 | 4838-5170 | - | Passenger Gene | Antibiotic Resistance |
| NAD(P)H-dependent oxidoreductase | Tn6007 | 5364-5879 | - | Passenger Gene | Other |
| intI1 | Tn6007 | 6040-7053 | + | Integron Integrase | Class 1 |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniA | TniA | Tn6007 | 559 | 144-1823 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | Tn402-like || homologous to TnsB of Tn7 || original is nonfunctional due to 1 bp deletion | ||||||||||
| Protein Sequence: | MASDTLPIAE QGVATLPDAA WAQARHRTEI IGPLAALEVV GHEAADAAAQ ALGLSRRQVY VLIRRARQGA GFVTDLVPGQ SGGGKGKGRL PESVERIIRE LLQKRFLTKQ KRSLAAFHRE VAQACKAQKL RVPARNTLAL RIAGLDPLKA TRRREGQDAS RSLQGVGGEP PAVTAPLEQV QIDHTVIDLI VVDERDRQPI GRPYLTIAID VFTRCVLGMV VTLEAPSSVS VGLCLVHVAC DKRPWLEGLN IEMDWPMSGK PRLLYLDNAA EFKSEALRRG CEQHGIRLDY RPPGQPHYGG IVERIIGTAM QMIHDELPGT TFSNPDQRGD YDSENKAALT LRELERWLTL AVGTYHGSVH NGLLQPPAAR WSEAVARVGV PAVVTRALAF LVDFLPIIRR TLTRTGFVID HIHYYADALK PWIARRDRLP AFLIRRDPRD ISRIWVLEPE GQHYLEIPYR TLSHPAVTLW EQRQALTKLR QQGREQVDES ALFRMIGQMR EIVTTAQKAT RKARRDADRR QHLKASPPPD KPIPPKTDVA DPQADNLPPA KPFDQIEEW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniB | TniB | Tn6007 | 302 | 1826-2734 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Comment: | homologous to TnsC protein of Tn7 putative ATP-binding protein | ||||||||||
| Protein Sequence: | MDEYPIIDLS HLLPAAQGLA RLPADERIQR LRADRWIGYP RAVEALNRLE TLYAWPNKQR MPNLLLVGPT NNGKSMIIEK FRRTHPASSD ADQEHMPVLV VQMPSEPSVI RFYVALLAAM GAPLRPRPRL PEMEQLALAL LRKVGVRMLV IDELHNVLAG NSVNRREFLN LLRFLGNELR IPLVGVGTRD AYLAIRSDDQ LENRFEPMML PVWEANDDCC SLLASFAASL PLRRPSSIAT LDMARYLLTR SEGTIGELAH LLMAAALVAV ESGEEAINHR TLSMADYTGP SERRRQFERE LM |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniQ | TniQ | Tn6007 | 405 | 2731-3948 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Target Site Selection | ||||||||||
| Comment: | Tn402-like || Protein: ACE81793.1 | ||||||||||
| Protein Sequence: | VKPAPRWPLH PAPKEGEALS SWLNRVALCY HMEVSDLLEH DLGHGQVDDL DTAPPLSLLM MLFQRSGIEL DRLRCMSFAG WVPWLLDSLD DQIPDALETY AFQLSVLLPK LRRRTRSITN WRAWLPSQPI HRACPLCLND PANQAVLLAW KLPLMLSCPL HGCWLESYWG VPGRFLGWDN ADTAPRTASD AIAVMDRRTW QALTTGHVEL PRRRIHAGLW FRLIRTLLDE LNTPLSTCGT CAGYLRQVWE GCGHPLRAGQ SLWRPYETLN PAVRLQMLEA AATAISLIEV RDISPPGEHA KLFWSEPQTG FTSGLPAKAL KPEPVDHWQR AVKAIDDAII EARHDPETAR SLFALASYGR RDPASLEQLR ATFAKEGIPT EFLSHYEPDE PFACLRQNDG LSDKF |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniR | TniR | Tn6007 | 207 | 4010-4633 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Comment: | resolution of cointegrates || Protein: ACE81792.1 || identical to tniR (Tn1721) | ||||||||||
| Protein Sequence: | MLIGYMRVSK ADGSQSTNLQ RDALIAAGVS LAHLYEDLAS GRRDDRPGLA ACLKALREGD TLIVWKLDRL GRDLRHLINT VHDLTARSVG LKVLTGHGAA VDTTTAAGKL VFGIFAALAE FERELISERT VAGLISARAR GRKGGRPFKM TAAKLRLAMA SMGQPETKVG DLCEELGITR QTLYRHVSPK GELRPDGVKL LSLGSAA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | qacL (ARO:3005098) | QacL | Tn6007 | 110 | 4838-5170 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic efflux (ARO:0010000) | ||||||||||
| Sequence Family: | small multidrug resistance (SMR) antibiotic efflux pump (ARO:0010003) | ||||||||||
| Target: | quaternary ammonium salts | ||||||||||
| Comment: | subunit of the qac multidrug efflux pump||loose match to reference sequence for ARO:3005098 (bitscore:173) | ||||||||||
| Protein Sequence: | MKNWLFLATA IIFEVIATSA LKSSEGFTRL VPSFIVVAGY AAAFYFLSLT LKSIPVGIAY AVWSGLGIVL VTAIAWVLHG QKLDMWGFVG VGFIISGVAV LNLLSKASVH |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | NAD(P)H-dependent oxidoreductase | NAD(P)H-dependent oxidoreductase | Tn6007 | 171 | 5364-5879 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Protein Sequence: | MSKTIAVFAS ARRNGNTGRL IDWISSELDI GVINLLDKDI SPYDYDHKNI GDDFLSVMNQ LLDYENIILA TPVYWYGPSA QMKVFIDRTS DFLDVDELKD IGRRLRSKTG FVVCTSISSD ADSSFLNSFK DTFRYLGMGY GGYVHANCEN GFNSQDYQAD VDRFIHLVKN N |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | intI1 | IntI1 | Tn6007 | 337 | 6040-7053 | + |
|---|
| Class: | Integron Integrase | ||||||||||
| Subclass: | Class 1 | ||||||||||
| Sequence Family: | Class 1 Integron Tyrosine Integrase | ||||||||||
| Transposase Chemistry: | Tyrosine | ||||||||||
| Protein Sequence: | MKTATAPLPP LRSVKVLDQL RERIRYLHYS LRTEQAYVHW VRAFIRFHGV RHPATLGSSE VEAFLSWLAN ERKVSVSTHR QALAALLFFY GKVLCTDLPW LQEIGRPRPS RRLPVVLTPD EVVRILGFLE GEHRLFAQLL YGTGMRISEG LQLRVKDLDF DHGTIIVREG KGSKDRALML PESLAPSLRE QLSRARAWWL KDQAEGRSGV ALPDALERKY PRAGHSWPWF WVFAQHTHST DPRSGVVRRH HMYDQTFQRA FKRAVEQAGI TKPATPHTLR HSFATALLRS GYDIRTVQDL LGHSDVSTTM IYTHVLKVGG AGVRSPLDAL PPLTSER |
||||||||||
1. Labbate M, Roy Chowdhury P, Stokes HW.
A class 1 integron present in a human commensal has a hybrid transposition module compared to Tn402: evidence of interaction with mobile DNA from natural environments.. J Bacteriol. 2008 Aug;190(15):5318-27. doi: 10.1128/JB.00199-08. Epub 2008 May 23. PubMed ID:
18502858
.
2. Ghaly TM, Chow L, Asher AJ, Waldron LS, Gillings MR. Evolution of class 1 integrons: Mobilization and dispersal via food-borne bacteria.. PLoS One. 2017 Jun 6;12(6):e0179169. doi: 10.1371/journal.pone.0179169. eCollection 2017. PubMed ID: 28586403 .
2. Ghaly TM, Chow L, Asher AJ, Waldron LS, Gillings MR. Evolution of class 1 integrons: Mobilization and dispersal via food-borne bacteria.. PLoS One. 2017 Jun 6;12(6):e0179169. doi: 10.1371/journal.pone.0179169. eCollection 2017. PubMed ID: 28586403 .