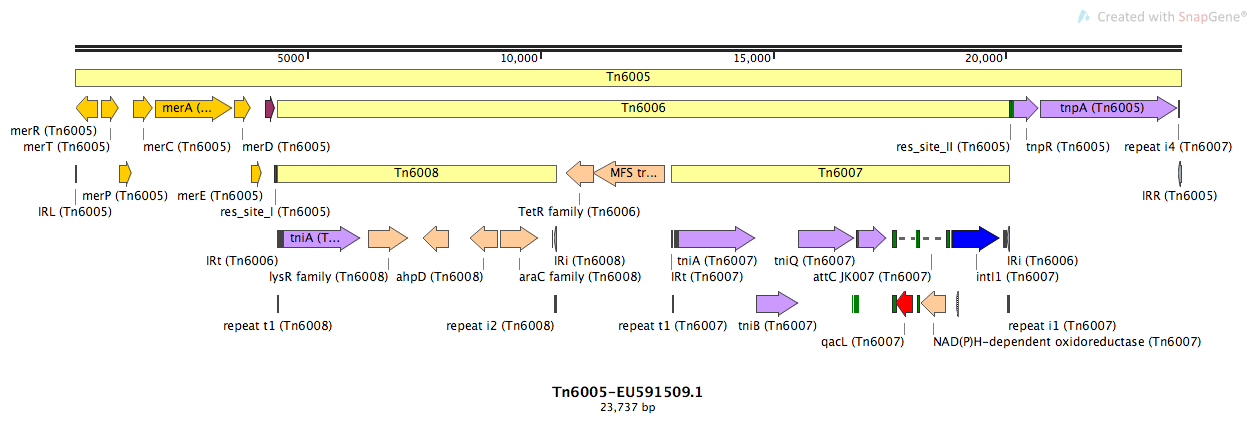
Mobile element type:
Transposon
Name:
Tn6005
Synonyms:
Accession:
Tn6005-EU591509.1
Family:
Tn3
Group:
Tn21
First isolate:
ND
Partial:
ND
Evidence of transposition:
Yes
Host Organism:
Enterobacter cloacaeJKB7
Date of Isolation:
None
Country:
None
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-38 | + | 38 | GGGGTCGTCT CAGAATTCGG AAAATAAAGC ACGCTAAG |
| IRR | 23697-23737 | - | 41 | GGGGTCGTCT CAGAAAACGG AAAATAAAGC ACGCTAAGCC G |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res_site_I | Tn6005 | 4295-4325 | + | 31 | CGTCAGATTG AGGCATACCC TAACTTGATG T |
| repeat t3 | Tn6007 | 12897-12916 | + | 20 | TCAGAAGTCA TCTGCACCAA |
| r4 | Tn6007 | 16696-16709 | - | 14 | TCGTAATGTG ACAA |
| r3 | Tn6007 | 16747-16760 | + | 14 | GGGTTAAGTG ACAA |
| res | Tn6007 | 16788-16822 | + | 35 | ACACTGTCAC ATAATCGAAC GTATACGTGA CGGGT |
| r2 | Tn6007 | 16791-16804 | - | 14 | CGATTATGTG ACAG |
| r1 | Tn6007 | 16807-16820 | + | 14 | CGTATACGTG ACGG |
| attC qacL (Tn6007) | Tn6007 | 17555-17647 , 18087-18092 | + | 99 | CGTTTGACGT GAGGGGCCGC CGTAGCGGCG AAGCCGCGAA GGGAACCCGC AAGCGCAGCT TGTGGGCGGT CCCTCTCGAC GGAATGGTTA GATATCTAA |
| attC qacL core | Tn6007 | 17555-17647 | + | 93 | CGTTTGACGT GAGGGGCCGC CGTAGCGGCG AAGCCGCGAA GGGAACCCGC AAGCGCAGCT TGTGGGCGGT CCCTCTCGAC GGAATGGTTA GAT |
| attC JK007 | Tn6007 | 18093-18146 , 18717-18722 | + | 60 | CGCTGGAGTT AAGCCGCGGC GCGTAGCGCC GTCGGCTTGA ACGACTTGTT ATACGTATAA |
| attC JK007 core | Tn6007 | 18093-18146 | + | 54 | CGCTGGAGTT AAGCCGCGGC GCGTAGCGCC GTCGGCTTGA ACGACTTGTT ATAC |
| attI | Tn6007 | 18723-18778 | + | 56 | CTTTGTTTTA GGGCGACTGC CCTGCTGCGT AACATCGTTG CTGCTCCATA ACATCA |
| res_site_II | Tn6005 | 20083-20117 | + | 35 | TGTCAGAATA GAGTTAAATT TCCTATTGAT TGACA |
| res_site_III | Tn6005 | 20120-20151 | + | 32 | TTCCGTCAAA GGTAATAGAT TTCATCCTGA CA |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| merR | Tn6005 | 34-489 | - | Passenger Gene | Heavy Metal Resistance |
| merT | Tn6005 | 561-962 | + | Passenger Gene | Heavy Metal Resistance |
| merP | Tn6005 | 959-1234 | + | Passenger Gene | Heavy Metal Resistance |
| merC | Tn6005 | 1262-1687 | + | Passenger Gene | Heavy Metal Resistance |
| merA | Tn6005 | 1726-3411 | + | Passenger Gene | Heavy Metal Resistance |
| merD | Tn6005 | 3429-3794 | + | Passenger Gene | Heavy Metal Resistance |
| merE | Tn6005 | 3791-4027 | + | Passenger Gene | Heavy Metal Resistance |
| urf-2Y-WP_000993245.1 | Tn6005 | 4093-4305 | + | Passenger Gene | Hypothetical |
| tniA | Tn6008 | 4486-6162 | + | Transposase | |
| lysR family | Tn6008 | 6309-7193 | + | Passenger Gene | Other |
| uracil-DNA glycosylase family | Tn6008 | 7493-8023 | - | Passenger Gene | Other |
| ahpD | Tn6008 | 8495-9070 | - | Passenger Gene | Other |
| araC family | Tn6008 | 9134-9988 | + | Passenger Gene | Other |
| TetR family | Tn6006 | 10567-11133 | - | Passenger Gene | Other |
| MFS transporter | Tn6006 | 11144-12655 | - | Passenger Gene | Other |
| tniA | Tn6007 | 12963-14642 | + | Transposase | |
| tniB | Tn6007 | 14645-15553 | + | Accessory Gene | |
| tniQ | Tn6007 | 15550-16767 | + | Accessory Gene | Target Site Selection |
| tniR | Tn6007 | 16829-17452 | + | Accessory Gene | Resolvase |
| qacL (ARO:3005098) | Tn6007 | 17657-17989 | - | Passenger Gene | Antibiotic Resistance |
| NAD(P)H-dependent oxidoreductase | Tn6007 | 18183-18698 | - | Passenger Gene | Other |
| intI1 | Tn6007 | 18859-19872 | + | Integron Integrase | Class 1 |
| tnpR | Tn6005 | 20172-20729 | + | Accessory Gene | Resolvase |
| tnpA | Tn6005 | 20732-23704 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merR | MerR | Tn6005 | 151 | 34-489 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MQINFENLTI GVFAKAAGVN VETIRFYQRK GLLPEPDKPY GSIRRYGEAD VTRVRFVKSA QRLGFSLDEI AELLRLEDGT HCEEASGLAE HKLKDVREKM ADLARMEAVL SELVCACHAR KGNVSCPLIA SLQDGTKLAA SARGSHGVTT P |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merT | MerT | Tn6005 | 133 | 561-962 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | response to mercury ion (GO:0046689) | ||||||||||
| Target: | Mercury | ||||||||||
| Comment: | ProteinID:ACE81807.1 | ||||||||||
| Protein Sequence: | MSEPQKSEPQ KSEPQNGRGA LFAGGLAAIL ASACCLGPLV LIALGFSGAW IGNLTVLEPY RPIFIGAALV ALFFAWRRIY RPAQACNPGE VCAISPRCEV LTSSFSGSWP RWSWSRSDFP TSCHFSINHR SSS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merP | MerP | Tn6005 | 91 | 959-1234 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MKKLFAALAL AAVVAPVWAA TQTVTLSVPG MTCASCPITV KHALSKVEGV SKTDVSFDKR QAVVTFDDAK TNVQKLTKAT EDAGYPSSLK R |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merC | MerC | Tn6005 | 141 | 1262-1687 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MGLITRIAGK TGALGSVVSA MGCAACFPAI ASFGAAIGLG FLSQYEGLFI GILLPMFAGI ALLANAIAWL NHRQWRRTAL GTIGPILVLA AVFLMRAYGW QSGGLLYVGL ALMVGVSVWD FISPAHRRCG PDSCELPEQR G |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merA | MerA | Tn6005 | 561 | 1726-3411 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MTTLKITGMT CDSCAAHVKE ALEKVPGVQS ALVSYPKGTA QLAIEAGTSS DALTTAVAGL GYEATLADAP PTDNRAGLLD KMRGWIGAAD KPSGNERPLQ VVVIGSGGAA MAAALKAVEQ GAQVTLIERG TIGGTCVNVG CVPSKIMIRA AHIAHLRRES PFDGGMPPTP PTILRERLLA QQQARVEELR HAKYEGILDG NSAITVLHGE ARFKDDQSLI VSLNEGGERV VMFDRCLVAT GASPAVPPIP GLKESPYWTS TEALASDTIP ERLAVIGSSV VALELAQAFA RLGSKVTALA RNTLFFREDP AIGEAVTAAF RAEGIEVLEH TQASQVAHMD GEFVLTTTHG ELRADKLLVA TGRTPNTRSL ALEAAGVAVN AQGAIVIDKG MRTSSPNIYA AGDCTDQPQF VYVAAAAGTR AAINMTGGDA ALDLTAMPAV VFTDPQVATV GYSEAEAHHD GIETDSRLLT LDNVPRALAN FDTRGFIKLV IEEGSGRLIG VQAVAPEAGE LIQTAVLAIR NRMTVQELAD QLFPYLTMVE GLKLAAQTFS KDVKQLSCCA G |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merD | MerD | Tn6005 | 121 | 3429-3794 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MSAYTVSRLA LDAGVSVHIV RDYLLRGLLR PVAYTTGGYG LFDDTALQRL RFVRAAFEAG IGLDALARLC RALDAADGDG ASAQLAVLRQ LVERRREALA SLEMQLAAMP TEPAQHAESL P |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merE | MerE | Tn6005 | 78 | 3791-4027 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MNSPEHLPSE THKPITGYLW GALAVLTCPC HLPILAIVLA GTTAGAFIGE HWGIAALTLT GLFVLSVTRL LRAFKGRS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | urf-2Y-WP_000993245.1 | Urf-2Y-WP_000993245.1 | Tn6005 | 70 | 4093-4305 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Hypothetical | ||||||||||
| Protein Sequence: | MNANAPNTAS CTTCCVCCKE IPLDAAFTPE GAEYVEHFCG LDCYERFQAR AKAATESDIA PVPGGSQPSD |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniA | TniA | Tn6008 | 559 | 4486-6162 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | homologous to TnsB of Tn7 | ||||||||||
| Protein Sequence: | MTSDTPPIAA QGVATLPDEA WAQARHRTEI IGPLAALEVV GHEAADEAAQ ALGLSRRQVY VLIRRARQGT GLVTDLTPGR SGGGKGKGRL PEPVERIIRE LLQKRFLTKQ KRSLAAFHRE VAQACKTQKL PVPARNTVAQ RIAGLHPAKI ARSRGGQDAA RPLQGAGGIP PEVTMPLEQV QIDHTVIDLI VVDERDRQPI GRPYLTLAID VFTRCVLGMV VTLEAPSAVS VGLCLAHAAC DKRPWLEGLN VEMDWPMSGK PRLLYLDNAA EFKSEALRRG CEQHGIRLDY RPPGQPHYGG IVERIIGTAM QMIHDELPGT TFSNPGQRGE YDSEKMATLT LRELERWLAL AVGTYHGSVH NGLLQPPAAR WAEAVERVGV PAVVTRPTAF LVDFLPVIRR TLTRTGFVID HIHYYADALK PWIARRERLP AFLIRRDPRD ISRIWVLEPE GQHYLEIHYR TLSHPAVTLW EQRQALAKLR QLGREQVDES ALFRMIGQMR EIVTTAQKAT RKARRDADRR QHLKTSEPPA KPIPPDVDMA DPQADNLPPA KPFDQIEEW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | lysR family | LysR family | Tn6008 | 294 | 6309-7193 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Comment: | putative transcriptional regulator, proteinID: ACA14445.1 | ||||||||||
| Protein Sequence: | MREISLDRLR TLVAIADLGS FAEAARVLHL APPTVSLHIA DLESRVGGKL LSRTRGRIQP SAIGETLVER ARRLLADAEQ ALEDVERQVQ GLAGRVRLGA STGAIAQLMP QALETLGQRH PAIDVQVAVL TSQETLKKLA EGSLEIGLVA LPQTPVKELR IEPWRRDPVM AFLPARWECP DVVTPGWLAA QPLILNDKTT RLSRLTSEWF ASDGRQPTPR IQLNYNDAIK SLVAAGYGAT LLPHEASTPL PDTRIVMRPL QPLLWRQLGI AHRGGDVERP TQHVLDVLWG LSAG |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | uracil-DNA glycosylase family | Uracil-DNA glycosylase family | Tn6008 | 176 | 7493-8023 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Protein Sequence: | MAPPSCASHP FRKRAMPTLD AFLTDVRACT RCAPHLPHGV QPVFQFHPSA PILIAGQAPG ARVHASGVPF DDASGARLRA WLGVDRDTFY DPTRIAILPM GFCYPGTGKS GDLPLRPECA PRLARSLHAA FRAPVAGHRA GQLRHGLPPR YWQDAAHPGG RSLARALAAS VPPASP |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | ahpD | AhpD | Tn6008 | 191 | 8495-9070 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Comment: | putative alkylhydroperoxidase || proteinID: ACA14447.1 | ||||||||||
| Protein Sequence: | MPTCSCTSPT TEIDMTQIAP LTIDTADAAT AATLKAVKAK LGMVPNLFAT LAHAPAALNG YLGLSETLGT GRLNASQREI VALAAAQANR CQYCLSAHTL IGKGAGLSAE AIAAARTGQA ANALDDAIAG FARALVEQRG VVSADAMANY RRAGLDDGLI LEVIANVALN TLTNYTNHIA DPTVDFPVVA V |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | araC family | AraC family | Tn6008 | 284 | 9134-9988 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Protein Sequence: | MESGMADRLA GLLRHYSLSA RVFHSGAFCG QHHYAAPHGY IHLVRRGPIT ARSPVHEDLL VTEPSLLFYP RVASHRFVAA PGDTAEQLCA EVDLGASTGN PLAMALPSML LIPLADLPGL GPTLELLFAE AERDQCGRQA AIDRLCELLL IQLLRYLMDG RLGATGLLAG LADPKLARAI TAMHDAPQTA WSLEALAAKA GMSRARFAAA FKDAVGVTPG DYLADWRMNV SCTLLKQGRP VAVVADRVGY GSPNALARAF RVRMGCAPRD WLAQQRGDAA LSGS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | TetR family | TetR family | Tn6006 | 188 | 10567-11133 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Protein Sequence: | MARASKRLEV LEAIVSIIER DGLTAVTLDA VALETGMTRA GLLYHFPSRE ALILATHEHL TRSWEQELEA SAGNTSDRAT DAERHAAYIN TCARAARRVE LLLILESSDN RQLGDLWQQV IDRWAPPAPI GNDAAELDRF ISRLAADGLW IHEALSSRPL PEQLRKRIAA RLVAMAAGPV EAEENLPK |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | MFS transporter | MFS transporter | Tn6006 | 503 | 11144-12655 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Comment: | major facilitator superfamily-related protein || ProteinID: ACE81795.1 | ||||||||||
| Protein Sequence: | MPAAHSNRWV LLVTVAAGLL LIVLDNSVLY TALPTLTREL GATATQGLWI INAYPLVMAG LLLGTGTLGD RIGHRRMFLI GLVLFGVASI VAAYSPTAEI LIGARAFLAV GAAAMMPATL ALIRVTFEDD RERNIAIAIW GSLSVVGAAL GPIIGGFLLG HFWWGSVFLI NVPVVVAAFI SALIVAPKVA GDATKPWDVV SSFQALVALS AFVIAIKESA HAGQSWAVPA ISLLVAILAG ALFVRRQLRL PFPLLDFSIF RNAAFTSGVL AAAFSLFAIG GVELATTQRF QLVAGFTPLE AGMLVSAAAL GSLPTALLGG AFLHRIGLRI LIAGGLAAGS LAVLLATWGI THGLGWLIAG LALTGAGVGA TMSVASTAIV GNVPVHRAGM ASSVEEVSYE FGSLFAVTIL GSLLAYLYTV NVVFPAGTSE AARDSMASAL VFANEAGADG VVVRQAAGIA FDHAYTVVMY VAAGVLAVGA LITGILLRRY GPGSQSSAYP TQH |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniA | TniA | Tn6007 | 559 | 12963-14642 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | Tn402-like || homologous to TnsB of Tn7 || original is nonfunctional due to 1 bp deletion | ||||||||||
| Protein Sequence: | MASDTLPIAE QGVATLPDAA WAQARHRTEI IGPLAALEVV GHEAADAAAQ ALGLSRRQVY VLIRRARQGA GFVTDLVPGQ SGGGKGKGRL PESVERIIRE LLQKRFLTKQ KRSLAAFHRE VAQACKAQKL RVPARNTLAL RIAGLDPLKA TRRREGQDAS RSLQGVGGEP PAVTAPLEQV QIDHTVIDLI VVDERDRQPI GRPYLTIAID VFTRCVLGMV VTLEAPSSVS VGLCLVHVAC DKRPWLEGLN IEMDWPMSGK PRLLYLDNAA EFKSEALRRG CEQHGIRLDY RPPGQPHYGG IVERIIGTAM QMIHDELPGT TFSNPDQRGD YDSENKAALT LRELERWLTL AVGTYHGSVH NGLLQPPAAR WSEAVARVGV PAVVTRALAF LVDFLPIIRR TLTRTGFVID HIHYYADALK PWIARRDRLP AFLIRRDPRD ISRIWVLEPE GQHYLEIPYR TLSHPAVTLW EQRQALTKLR QQGREQVDES ALFRMIGQMR EIVTTAQKAT RKARRDADRR QHLKASPPPD KPIPPKTDVA DPQADNLPPA KPFDQIEEW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniB | TniB | Tn6007 | 302 | 14645-15553 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Comment: | homologous to TnsC protein of Tn7 putative ATP-binding protein | ||||||||||
| Protein Sequence: | MDEYPIIDLS HLLPAAQGLA RLPADERIQR LRADRWIGYP RAVEALNRLE TLYAWPNKQR MPNLLLVGPT NNGKSMIIEK FRRTHPASSD ADQEHMPVLV VQMPSEPSVI RFYVALLAAM GAPLRPRPRL PEMEQLALAL LRKVGVRMLV IDELHNVLAG NSVNRREFLN LLRFLGNELR IPLVGVGTRD AYLAIRSDDQ LENRFEPMML PVWEANDDCC SLLASFAASL PLRRPSSIAT LDMARYLLTR SEGTIGELAH LLMAAALVAV ESGEEAINHR TLSMADYTGP SERRRQFERE LM |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniQ | TniQ | Tn6007 | 405 | 15550-16767 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Target Site Selection | ||||||||||
| Comment: | Tn402-like || Protein: ACE81793.1 | ||||||||||
| Protein Sequence: | VKPAPRWPLH PAPKEGEALS SWLNRVALCY HMEVSDLLEH DLGHGQVDDL DTAPPLSLLM MLFQRSGIEL DRLRCMSFAG WVPWLLDSLD DQIPDALETY AFQLSVLLPK LRRRTRSITN WRAWLPSQPI HRACPLCLND PANQAVLLAW KLPLMLSCPL HGCWLESYWG VPGRFLGWDN ADTAPRTASD AIAVMDRRTW QALTTGHVEL PRRRIHAGLW FRLIRTLLDE LNTPLSTCGT CAGYLRQVWE GCGHPLRAGQ SLWRPYETLN PAVRLQMLEA AATAISLIEV RDISPPGEHA KLFWSEPQTG FTSGLPAKAL KPEPVDHWQR AVKAIDDAII EARHDPETAR SLFALASYGR RDPASLEQLR ATFAKEGIPT EFLSHYEPDE PFACLRQNDG LSDKF |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniR | TniR | Tn6007 | 207 | 16829-17452 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Comment: | resolution of cointegrates || Protein: ACE81792.1 || identical to tniR (Tn1721) | ||||||||||
| Protein Sequence: | MLIGYMRVSK ADGSQSTNLQ RDALIAAGVS LAHLYEDLAS GRRDDRPGLA ACLKALREGD TLIVWKLDRL GRDLRHLINT VHDLTARSVG LKVLTGHGAA VDTTTAAGKL VFGIFAALAE FERELISERT VAGLISARAR GRKGGRPFKM TAAKLRLAMA SMGQPETKVG DLCEELGITR QTLYRHVSPK GELRPDGVKL LSLGSAA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | qacL (ARO:3005098) | QacL | Tn6007 | 110 | 17657-17989 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic efflux (ARO:0010000) | ||||||||||
| Sequence Family: | small multidrug resistance (SMR) antibiotic efflux pump (ARO:0010003) | ||||||||||
| Target: | quaternary ammonium salts | ||||||||||
| Comment: | subunit of the qac multidrug efflux pump||loose match to reference sequence for ARO:3005098 (bitscore:173) | ||||||||||
| Protein Sequence: | MKNWLFLATA IIFEVIATSA LKSSEGFTRL VPSFIVVAGY AAAFYFLSLT LKSIPVGIAY AVWSGLGIVL VTAIAWVLHG QKLDMWGFVG VGFIISGVAV LNLLSKASVH |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | NAD(P)H-dependent oxidoreductase | NAD(P)H-dependent oxidoreductase | Tn6007 | 171 | 18183-18698 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Protein Sequence: | MSKTIAVFAS ARRNGNTGRL IDWISSELDI GVINLLDKDI SPYDYDHKNI GDDFLSVMNQ LLDYENIILA TPVYWYGPSA QMKVFIDRTS DFLDVDELKD IGRRLRSKTG FVVCTSISSD ADSSFLNSFK DTFRYLGMGY GGYVHANCEN GFNSQDYQAD VDRFIHLVKN N |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | intI1 | IntI1 | Tn6007 | 337 | 18859-19872 | + |
|---|
| Class: | Integron Integrase | ||||||||||
| Subclass: | Class 1 | ||||||||||
| Sequence Family: | Class 1 Integron Tyrosine Integrase | ||||||||||
| Transposase Chemistry: | Tyrosine | ||||||||||
| Protein Sequence: | MKTATAPLPP LRSVKVLDQL RERIRYLHYS LRTEQAYVHW VRAFIRFHGV RHPATLGSSE VEAFLSWLAN ERKVSVSTHR QALAALLFFY GKVLCTDLPW LQEIGRPRPS RRLPVVLTPD EVVRILGFLE GEHRLFAQLL YGTGMRISEG LQLRVKDLDF DHGTIIVREG KGSKDRALML PESLAPSLRE QLSRARAWWL KDQAEGRSGV ALPDALERKY PRAGHSWPWF WVFAQHTHST DPRSGVVRRH HMYDQTFQRA FKRAVEQAGI TKPATPHTLR HSFATALLRS GYDIRTVQDL LGHSDVSTTM IYTHVLKVGG AGVRSPLDAL PPLTSER |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn6005 | 185 | 20172-20729 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Serine Site-SpecificRecombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MQGQRIGYVR VSSFDQNPER QLEGVQVARV FTDKASGKDT QRPELERLLA FVREGDTVVV HSMDRLARNL DDLRRIVQGL TQRGVRMEFV KEGLKFTGED SPMANLMLSV MGAFAEFERA LIRERQREGI VLAKQRGAYR GRKKSLNSEQ IAELKRRVAA GDQKTLVARD FGISRETLYQ YLRED |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn6005 | 990 | 20732-23704 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MPRRSILSAT ERESLLALPD AKDELIRHYM FNETDLSVIR QRRGAANRLG FAVQLCYLRF PGTFLGVDEP PFPPLLRMVA AQLKMPVESW SEYGQREQTR REHLVELQTV FGFKPFTMSH YRQAVHTLTE LALQTDKGIV LASALVENLR RQSIILPAMN AIERASAEAI TRANRRIYAA LTDSLLSPHR QRLDELLKRK DGSKVTWLAW LRQSPAKPNS RHMLEHIERL KSWQALDLPA GIERQVHQNR LLKIAREGGQ MTPADLAKFE VQRRYATLVA LAIEGMATVT DEIIDLHDRI IGKLFNAAKN KHQQQFQASG KAINDKVRMY GRIGQALIEA KQSGSDPFAA IEAVMPWDTF AASVTEAQTL ARPADFDFLH HIGESYATLR RYAPQFLGVL KLRAAPAAKG VLDAIDMLRG MNSDSARKVP ADAPTAFIKP RWAKLVLTDD GIDRRYYELC ALSELKNALR SGDVWVQGSR QFKDFDEYLV PVEKFATLKL ASELPLAVAT DCDQYLHDRL ELLEAQLATV NRMAAANDLP DAIITTASGL KITPLDAAVP DAAQAMIDQT AMLLPHLKIT ELLMEVDEWT GFTRHFTHLK TSDTAKDKTL LLTTILADAI NLGLTKMAES CPGTTYAKLS WLQAWHIRDE TYSTALAELV NAQFRQPFAG NWGDGTTSSS DGQNFRTGSK AESTGHINPK YGSSPGRTFY THISDQYAPF SAKVVNVGIR DSTYVLDGLL YHESDLRIEE HYTDTAGFTD HVFGLMHLLG FRFAPRIRDL GETKLFIPKG DAAYDALKPM ISSDRLNIKQ IRAHWDEILR LATSIKQGTV TASLMLRKLG SYPRQNGLAV ALRELGRIER TLFILDWLQS VELRRRVHAG LNKGEARNAL ARAVFFYRLG EIRDRSFEQQ RYRASGLNLV TAAIVLWNTV YLERATSALR GNGTALDDTL LQYLSPLGWE HINLTGDYLW RSSAKVGAGK FRPLRPLPPA |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| Tn6006-EU591509.1 | Tn6006 | transposon | 4343-20074 | + | 15732 |
| Tn6008-EU316185.1 | Tn6008 | transposon | 4343-10341 | + | 5999 |
| Tn6007-EU591509.1 | Tn6007 | transposon | 12820-20074 | + | 7255 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRt | Tn6006 | 4343-4367 | + | 25 | TGTCGTTTTC AGAAGACGAC CGCAC |
| repeat t1 | Tn6008 | 4351-4369 | + | 19 | TCAGAAGACG ACCGCACCA |
| repeat t2 | Tn6008 | 4391-4409 | - | 19 | TCAGGAGCCG TATGCACAC |
| repeat t3 | Tn6008 | 4420-4438 | + | 19 | TCAGGAGTCG TCTGCACCA |
| repeat t4 | Tn6008 | 4452-4470 | + | 19 | TCAATACTCG TGTGCACCA |
| repeat i3 | Tn6008 | 10250-10268 | - | 19 | TCAGGCAACG ACGGGCTGC |
| repeat i2 | Tn6008 | 10292-10310 | - | 19 | TCAGAAGCCG ACTGCACTA |
| IRi | Tn6008 | 10309-10341 | - | 33 | TGTCGTTTTC AGAAGACGGC TGCACTGAAC GTC |
| repeat i1 | Tn6008 | 10315-10333 | - | 19 | TCAGAAGACG GCTGCACTG |
| IRt | Tn6007 | 12820-12844 | + | 25 | TGTCATTTTC AGAAGACGAC TGCAC |
| repeat t1 | Tn6007 | 12828-12846 | + | 19 | TCAGAAGACG ACTGCACCA |
| repeat t2 | Tn6007 | 12869-12886 | - | 18 | TCAGGAGCCG ACTGCACA |
| repeat t4 | Tn6007 | 12929-12947 | + | 19 | TCAATACTCG TGTGCACCA |
| repeat i4 | Tn6007 | 19955-19973 | - | 19 | TCAGCGGACG CAGGGAGGA |
| repeat i3 | Tn6007 | 19983-20001 | - | 19 | TCAGGCAACG ACGGGCTGC |
| repeat i2 | Tn6007 | 20025-20043 | - | 19 | TCAGAAGCCG ACTGCACTA |
| IRi | Tn6006 | 20042-20074 | - | 33 | TGTCGTTTTC AGAAGACGGC TGCACTGAAC GTC |
| repeat i1 | Tn6007 | 20048-20066 | - | 19 | TCAGAAGACG GCTGCACTG |
1. Labbate M, Roy Chowdhury P, Stokes HW.
A class 1 integron present in a human commensal has a hybrid transposition module compared to Tn402: evidence of interaction with mobile DNA from natural environments.. J Bacteriol. 2008 Aug;190(15):5318-27. doi: 10.1128/JB.00199-08. Epub 2008 May 23. PubMed ID:
18502858
.
2. Ghaly TM, Chow L, Asher AJ, Waldron LS, Gillings MR. Evolution of class 1 integrons: Mobilization and dispersal via food-borne bacteria.. PLoS One. 2017 Jun 6;12(6):e0179169. doi: 10.1371/journal.pone.0179169. eCollection 2017. PubMed ID: 28586403 .
2. Ghaly TM, Chow L, Asher AJ, Waldron LS, Gillings MR. Evolution of class 1 integrons: Mobilization and dispersal via food-borne bacteria.. PLoS One. 2017 Jun 6;12(6):e0179169. doi: 10.1371/journal.pone.0179169. eCollection 2017. PubMed ID: 28586403 .