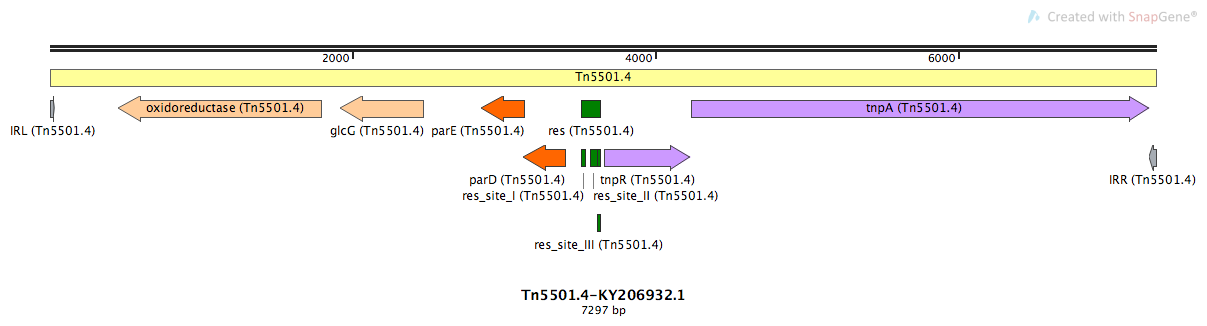
Mobile element type:
Transposon
Name:
Tn5501.4
Synonyms:
Accession:
Tn5501.4-KY206932.1
Family:
Tn3
Group:
Tn3000
First isolate:
Yes
Partial:
ND
Evidence of transposition:
ND
Host Organism:
Uncultured bacterium
Date of Isolation:
2016
Country:
Belgium
Molecular Source:
contig003 chromosome
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-38 | + | 38 | GGGGTTCTAA GCCGGAACCG CCGAAAATTC CGTCAGCC |
| IRR | 7260-7297 | - | 38 | GGGGTTCTAA GCCGGAACCG CCGAAATTTC CGTCATCC |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res | Tn5501.4 | 3508-3638 | + | 131 | ACCTGTCGGA AAACATTTGT TTTTCGACAG GCCTTCAACG GTCCTCTGCA CCAACCTCCG AGTGGCCGCA AAATTGTGCG GAAAACTCTG TCGCCAGACG CTACCATACG GAAACCTCGT CTTGATGGTT T |
| res_site_I | Tn5501.4 | 3508-3536 | + | 29 | ACCTGTCGGA AAACATTTGT TTTTCGACA |
| res_site_II | Tn5501.4 | 3570-3613 | + | 44 | TGGCCGCAAA ATTGTGCGGA AAACTCTGTC GCCAGACGCT ACCA |
| res_site_III | Tn5501.4 | 3614-3638 | + | 25 | TACGGAAACC TCGTCTTGAT GGTTT |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| oxidoreductase | Tn5501.4 | 456-1790 | - | Passenger Gene | Other |
| glcG | Tn5501.4 | 1922-2467 | - | Passenger Gene | Other |
| parE | Tn5501.4 | 2848-3132 | - | Passenger Gene | Toxin |
| parD | Tn5501.4 | 3129-3407 | - | Passenger Gene | Antitoxin |
| tnpR | Tn5501.4 | 3661-4239 | + | Accessory Gene | Resolvase |
| tnpA | Tn5501.4 | 4236-7265 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | oxidoreductase | Oxidoreductase | Tn5501.4 | 444 | 456-1790 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Protein Sequence: | MKRIKVSLLA VLLLLTALWL AADTLLPEPF TYFSFRTVFM QYSGVIGIGL MSVAMLLALR PKWLEPHLDG LDKMYRLHKW LGIAALVVAI VHWWWGKGTK WMVGWGWLEK PVRKPVAGET LGNIEGWLRS QRGFAESVGE WAFYAAAVLI VLALVKRFPY HWFVKTHKWI AVAYLALAYH SAVLTKVEYW TQPVGWLMAA LLLGGTVTAL LTLTGRIGTG RKVTGTIAGL IDYPALRVLE TAVVLEDGWR GHAAGQFAFI TSDRSEGAHP YTIASAWNPA DRRLVFITKA LGDHTSRLRE RLKIGMPVTV EGPYGCFNFE DAQPRQIWVG AGIGITPFIA RMKQRATTPD AKTIDLFHPT AEFEQAAIDR LTADAKAAGV RLHLLVDGKD GRLNGERIRA VIPEWQSASI WFCGPPRFGQ ALREDFLAHG LPPERFHQEL FQMR |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | glcG | GlcG | Tn5501.4 | 181 | 1922-2467 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Comment: | glc operon protein | ||||||||||
| Protein Sequence: | MRQILPNAHG MVCFCVALAI GAGFPTQEAV AQSAPAMGTA KAQPEYVKTV REISLASAQR LLANGLEIAG QREMRLAIAV VDSAGNLLAF VRMDGASIVT IDVAVGKART AAFLKAPSKV FEDMINSGQP SMATTPGLLP LQGGIPVVFD GEVIGAVGVS GSSGENDQTV ATRIAESFAR F |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | parE | ParE | Tn5501.4 | 94 | 2848-3132 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Toxin | ||||||||||
| Sequence Family: | ParE_toxin (Pfam:PF05016) | ||||||||||
| Target: | DNA gyrase | ||||||||||
| Protein Sequence: | VRVVWTPEAQ QDRADVWDYI AADNPRAAAR MDEIFSDAAA RLIQHPMLGK PGKIPGTREL IPHESYRLVY QIDGETVWIL TLVHTARLWP PVRD |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | parD | ParD | Tn5501.4 | 92 | 3129-3407 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antitoxin | ||||||||||
| Sequence Family: | parD (PDB:4Q2U) | ||||||||||
| Comment: | RelB/ParD/CcdA/DinJ | ||||||||||
| Protein Sequence: | MSKQAVFTMK LEPELRAEFM AEAEAAHRPA SQVLRELMRE FVQRQRESRE YDEFLRRKVE AGRASMRAGL GRSNDEVEAE FAARRASVAS QA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn5501.4 | 192 | 3661-4239 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MRVSSDSDRQ STNLQRDALL AAGVDARHLF EDHASGAKDD RAGLARALEF VRPGDVLVVW KLDRLGRSLS HLLAIVTSLK EKQVAFRSLT ENLDTTTPSG EFLFQVFGAL AQYERALIQE RVVAGLAAAR KRGRIGGRPQ AITGEKLEAI VAALDGGMSK AAVCRNFGVK RTTLIETLAR VGWTGSRGAS SR |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn5501.4 | 1009 | 4236-7265 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MTTKSERLTV LSDAEQEALY GLPDFDDAQR LEYLALTETE LALASSRPGL HAQVYCILQI GYFKAKHAFF RFDWSEVEHD CAFVLSRYFH GESFEHKPIS KHEHYTQREW IADLFGYRPW AAEFLAQLAQ QAAQTVRRDV MPGFIAAELI VWLNEHKIIR PGYTTLQELV SEALSAERRR LAGLLSEVLD ESAKAALGRL LVRDDTLSQL AALKQDAKDF GWRQMARERE KRATLEPLHR IAKALLPKLG VSQQNLLYYA SLANFYTVHD LRNLKADQTY LYLLCYAWVR YRQLSDNLVD AMAYHMKQLE DESSAGAKQS FVAEQVRRQQ DTPQVGRLLS LYIDDSVPDP TPFGDVRQRA YKIMPRDTLQ TTAQRMSVKP VSKLALHWQA VDGLAERIRR HLRPLYVALD LAGTDPGSPW LVALAWAKDV FAKQQRLSQR PLAECPAATL PKRLRPYLLT FDADGKPTGL HADRYEFWLY RQVRKRFQSG ELYLDDSLQH RHFSDELVSL DEKAAVLAQI DIPFLRQPLD AQLDALATEL RAQWLAFNRE LKQGKLTHLE YDKDTQKLTW RKPKGENQKA REKAFYEQLP FCDVADVFRF VNGQCQFLSA LTPLQPRYAK KVADADSLMA VIIAQAMNHG NQVMARTSDI PYHVLESAYQ QYLRHATLHA ANDCISNAIA ALPIFPYYSF DLDALYGAVD GQKFGVERPT VKARHSRKYF GRGKGVVAYT LLCNHVPLNG YLIGAHDYEA HHVFDIWYRN TSDIVPTAIT GDMHSVNKAN FAILHWFGLR FEPRFTDLGD QLKELYSADD PALYDQCLIR PAGRIDRDLI VSEKPNLDQI VATLGLKEMT QGTLIRKLCT YTAPNPTRRA VFEFDKLIRS IYTLRYLRDP QLERNVHRSQ NRIESYHQLR STIAQVGGKK ELTGRTDIEI EISNQCARLI ANAVIFYNSA ILSRLLMKYE ASGNAKAHAL LTQISPAAWR HILLNGHYTF QSDGKMIDLD ALVAGLELG |
||||||||||
1. Dunon V, Bers K, Lavigne R, Top EM, Springael D.
Targeted metagenomics demonstrates the ecological role of IS1071 in bacterial community adaptation to pesticide degradation.. Environ Microbiol. 2018 Nov;20(11):4091-4111. doi: 10.1111/1462-2920.14404. Epub 2018 Oct 5. PubMed ID:
30207068
.