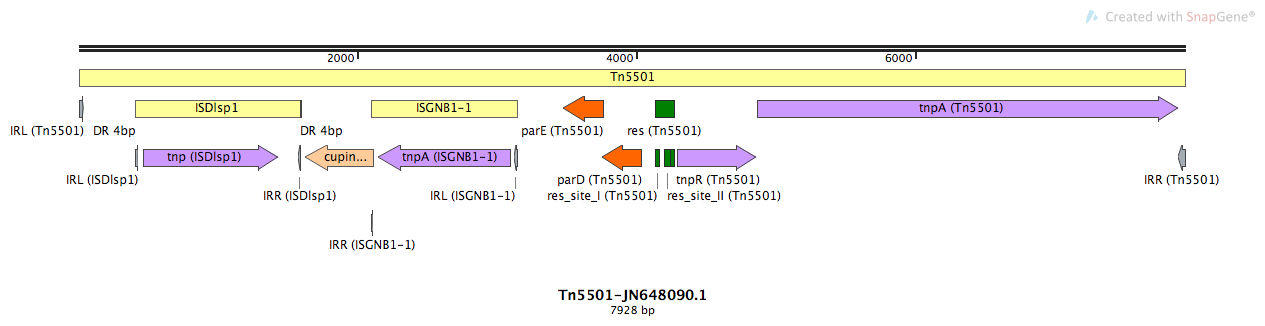
Mobile element type:
Transposon
Name:
Tn5501
Synonyms:
Accession:
Tn5501-JN648090.1
Family:
Tn3
Group:
Tn3000
First isolate:
Yes
Partial:
ND
Evidence of transposition:
Yes
Host Organism:
Delftia sp. KV29
Date of Isolation:
1998
Country:
Molecular Source:
plasmid pKV29
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-38 | + | 38 | GGGGTTCTAA GCCGGAACCG CCGAAAATTC CGTCAGCC |
| IRR | 7891-7928 | - | 38 | GGGGTTCTAA GCCAGAACCG CCGAAATTTC CGTCATCC |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res | Tn5501 | 4139-4269 | + | 131 | GCCTGTCGGA AAACATTTGT TTTTCGACAG GCCTTCAACG GTCCTCTGCA CCAACCTCCG AGTGGCCGCA AAATTGTGCG GAAAACTCTG TCGCCAGACG CTACCATACG GAAACCTCGT CTTAATGGTT T |
| res_site_I | Tn5501 | 4139-4167 | + | 29 | GCCTGTCGGA AAACATTTGT TTTTCGACA |
| res_site_II | Tn5501 | 4201-4244 | + | 44 | TGGCCGCAAA ATTGTGCGGA AAACTCTGTC GCCAGACGCT ACCA |
| res_site_III | Tn5501 | 4245-4269 | + | 25 | TACGGAAACC TCGTCTTAAT GGTTT |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| tnp | ISDlsp1 | 462-1436 | + | Transposase | |
| cupin2 | Tn5501 | 1626-2111 | - | Passenger Gene | Other |
| tnpA | ISGNB1-1 | 2150-3097 | - | Transposase | |
| parE | Tn5501 | 3479-3763 | - | Passenger Gene | Toxin |
| parD | Tn5501 | 3760-4038 | - | Passenger Gene | Antitoxin |
| tnpR | Tn5501 | 4292-4870 | + | Accessory Gene | Resolvase |
| tnpA | Tn5501 | 4867-7896 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnp | Tnp | ISDlsp1 | 324 | 462-1436 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MLTGMKQSSL ELNLSTRKTR KQELLAQMDR VVPWAALVEL IAPYYPEGKN GRPPFALEAM LRVHCMQQWF TLSDLAMEEA FFDTPIYREF AGLDAHGRMP DESTILRFRH RLEKHRLAEQ ILATVNDLLA ARGLLLKAGT AVDATLIAAP SSTKNKDRKR DPEMHSSQKG NEWHFGMKAH IGVDADSGLV HTVIGTSGNV ADVTEGNSLL HGEETDAFGD AGYQGAHKRP DARKDVTWHV AMRPGKRKEL DKENNPVDAL IDQVEKIKAS IRAKVEHPFR VIKRQFGYTK VRYRGLKKNT LQLKTLFALS NLWMVRHQLL GAQG |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | cupin2 | Cupin2 | Tn5501 | 161 | 1626-2111 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Comment: | Cupin 2conserved barrel domain protein | ||||||||||
| Protein Sequence: | MCNKAPQQRR VAPPFRAGYR QRRTLGYSCS VITSQAKLIF GVDMEMESRI FSVTEYIRPS DGEPIRSVVL ETKDSAVVVW HAHPGQEITA HVHPDGQDTW TVISGEAEYY QGGGKVAHLK AGDIAIAKPG QVHGALNTSP VPFVFVSVVA SGNAGFALAE K |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | ISGNB1-1 | 315 | 2150-3097 | - |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MSESKSAKPR YKTTNWAAYN AALKARGSLA IWLDKDMQWY APASGKRGRQ HVFSDAAIQF CLSIKCLFGL ALRQSLGLVQ SLLHLADLDW RVPDFSTVSR RQKTLQVQLP YRASTSALNL LVDSTGIKFL GEGEWKRKKH GAEYRRQWRK VHLGIDASTL EIRAIEVTDN SVGDAPMLPG LLGQIPPGEP IASVSTDGAY DTKACHAAIM ERGAQAIIPP RKNAQVWKSQ TPGAMVRNEA VLACKRLGWR IWKKWSGYHR RSLVETKMHC FKRLGERVMA RTFERQVTEL HVRVALLNRF TQLGCPTTVA VPAVA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | parE | ParE | Tn5501 | 94 | 3479-3763 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Toxin | ||||||||||
| Sequence Family: | ParE_toxin (Pfam:PF05016) | ||||||||||
| Target: | DNA gyrase | ||||||||||
| Protein Sequence: | VRVVWTPEAQ QDRADVWDYI AADNPRAAAR MDEIFSDAAA RLIQHPMLGK PGKIPGTREL IPHESYRLVY QIDGETVWIL TLVHTARLWP PVRD |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | parD | ParD | Tn5501 | 92 | 3760-4038 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antitoxin | ||||||||||
| Sequence Family: | parD (PDB:4Q2U) | ||||||||||
| Comment: | RelB/ParD/CcdA/DinJ | ||||||||||
| Protein Sequence: | MSKQAVFTMK LEPELRAEFM AEAEAAHRPA SQVLRELMRE FVQRQRESRE YDEFLRRKVE AGRASMRAGL GRSNDEVEAE FAARRASVAS QA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn5501 | 192 | 4292-4870 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MRVSSDSDRQ STNLQRDALL AVGVDARHLF EDHASGAKDD RAGLARALEF VRPGDVLVVW KLDRLGRSLS HLLAIVTSLK KKQVAFRSLT ENLDTTTPSG EFLFQVFGAL AQYERALIQE RVVAGLAAAR KRGRIGGRPQ AITGEKLEAI VAALDGGMSK AAVCRNFGVK RTTLIETLAR VGWTGSRGAS SR |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn5501 | 1009 | 4867-7896 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MTTKSERLTV LSDAEQEALY GLPDFDDAQR LEYLALTETE LALASSRPGL HAQVYCILQI GYFKAKHAFF RFDWSEVEHD CAFVLSRYFH GESFEHKPIS KHEHYTQREW IADLFGYRPW AAEFLAQLAQ QAAQTVRRDV MPGFIAAELI VWLNEHKIIR PGYTTLQELV SEALSAERRR LAGLLSEVLD ESAKAALGRL LVRDDTLSQL AALKQDAKDF GWRQMARERE KRATLEPLHR IAKALLPKLG VSQQNLLYYA SLANFYTVHD LRNLKADQTY LYLLCYAWVR YRQLSDNLVD AMAYHMKQLE DESSAGAKQS FVAEQVRRQQ DTPQVGRLLS LYIDDSVPDP TPFGDVRQRA YKIMPRDTLQ TTAQRMSVKP VSKLALHWQA VDGLAERIRR HLRPLYVALD LAGTDPGSPW LVALAWAKDV FAKQQRLSQR PLAECPAATL PKRLRPYLLT FDADGKPTDL HADRYEFWLY RQVRKRFQSG ELYLDDSLQH RHFSDELVSL DEKAAVLAQI DIPFLRQPLD AQLDALATEL RAQWLAFNRE LKQGKLTHLE YDKDTQKLTW RKPKGENQKA REKAFYEQLP FCDVADVFRF VNGQCQFLSA LTPLQPRYAK KVADADSLMA VIIAQAMNHG NQVMARTSDI PYHVLESAYQ QYLRHATLHA ANDCISNAIA ALPIFPYYSF DLDALYGAVD GQKFGVERPT VKARHSRKYF GRGKGVVAYT LLCNHVPLNG YLIGAHDYEA HHVFDIWYRN TSDIVPTAIT GDMHSVNKAN FAILHWFGLR FEPRFTDLGD QLKELYSADD PALYDQCLIR PAGRIDRDLI VSEKPNLDQI VATLGLKEMT QGTLIRKLCT YTAPNPTRRA VFEFDKLIRS IYTLRYLRDP QLERNVHRSQ NRIESYHQLR STIAQVGGKK ELTGRTDIEI EISNQCARLI ANAVIFYNSA ILSRLLMKYE ASGNAKAHAL LTQISPAAWR HILLNGHYTF QSDGKMIDLD ALVAGLELG |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| ISDlsp1-JN648090.1 | ISDlsp1 | insertion sequence | 409-1592 | + | 1184 |
| ISGNB1-1-EF628291 | ISGNB1-1 | insertion sequence | 2097-3143 | + | 1047 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRL | ISDlsp1 | 409-428 | + | 20 | GGAAATCCTG CAAAACCTCG |
| IRR | ISDlsp1 | 1574-1592 | - | 19 | GGATGTCCTG AACAACTCG |
| IRR | ISGNB1-1 | 2097-2111 | + | 15 | GGCTTTGTTG CACAA |
| IRL | ISGNB1-1 | 3129-3143 | - | 15 | GGCTCTGTTG CACAA |
1. Stolze Y, Eikmeyer F, Wibberg D, Brandis G, Karsten C, Krahn I, Schneiker-Bekel S, Viehover P, Barsch A, Keck M, Top EM, Niehaus K, Schluter A.
IncP-1beta plasmids of Comamonas sp. and Delftia sp. strains isolated from a wastewater treatment plant mediate resistance to and decolorization of the triphenylmethane dye crystal violet.. Microbiology (Reading). 2012 Aug;158(Pt 8):2060-2072. doi: 10.1099/mic.0.059220-0. Epub 2012 May 31. PubMed ID:
22653947
.