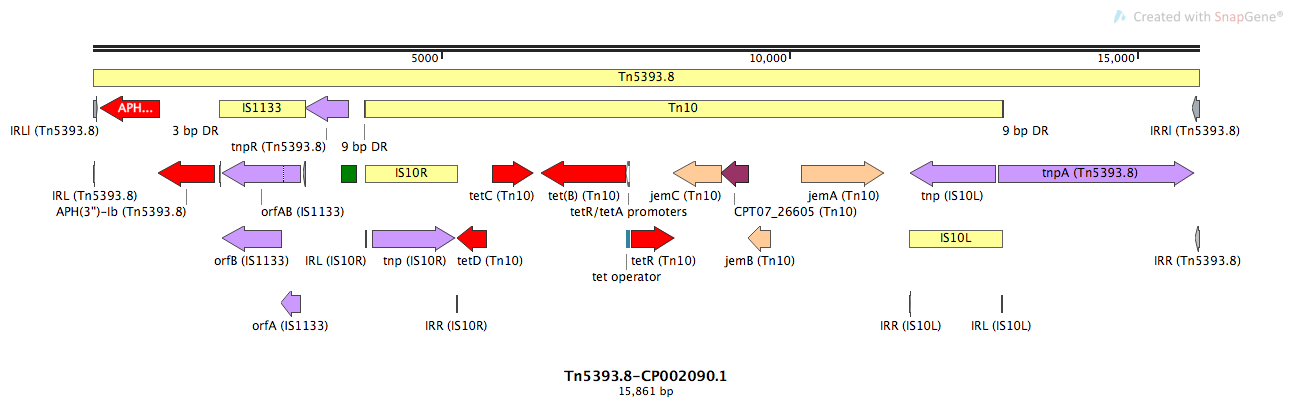
Mobile element type:
Transposon
Name:
Tn5393.8
Synonyms:
Accession:
Tn5393.8-CP002090.1
Family:
Tn3
Group:
Tn163
First isolate:
ND
Partial:
ND
Evidence of transposition:
ND
Host Organism:
Salmonella enterica subsp. enterica serovar Kentucky
Date of Isolation:
Country:
Molecular Source:
plasmid pCS0010A
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRLl | 1-81 | + | 81 | GGGGTCGTTT GCGGGAGAGG GCGAAATCCT ACGCTAAGGC TTTGGCCAAC GATATTCTCC GGTAAGATTG ATGTGTTCCC A |
| IRL | 1-40 | + | 40 | GGGGTCGTTT GCGGGAGAGG GCGAAATCCT ACGCTAAGGC |
| IRRl | 15781-15861 | - | 81 | GGGGTCGTTT GCGGGAGGGG GCGGAATCCT ACGCTAAGGC TTTGGCCAGC GATATTCTCC GGTGAGATTG ATGTGTTCCC A |
| IRR | 15822-15861 | - | 40 | GGGGTCGTTT GCGGGAGGGG GCGGAATCCT ACGCTAAGGC |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res | Tn5393.8 | 3565-3786 | + | 222 | TGTCCCGTTC GACACCTGCG GCGCGCAAGG CGTCGTGCTG CAGGTCGAGA GACTGCGAGC CATCGGCTTT GGAGACGCGG GCATATCCGA TCAGCATGTA TCACAAACGT TGGTTTGAGG CGGCGCTTCG GCCACGATTG CATTGACCTC TGGAAATGTA TCTCAACCAG CTTCATAAAC AAAGCGTCTT GAACGCTATC AGATTTTGAA AAAGGAACAT GT |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| APH(6)-Id (ARO:3002660) | Tn5393.8 | 107-952 | - | Passenger Gene | Antibiotic Resistance |
| APH(3'')-Ib (ARO:3002639) | Tn5393.8 | 943-1746 | - | Passenger Gene | Antibiotic Resistance |
| orfAB | IS1133 | 1853-2982 | - | Transposase | |
| orfB | IS1133 | 1853-2710 | - | Transposase | |
| orfA | IS1133 | 2707-2982 | - | Accessory Gene | Regulator |
| tnpR | Tn5393.8 | 3050-3661 | - | Accessory Gene | Resolvase |
| tnp | IS10R | 4016-5224 | + | Transposase | |
| tetD | Tn10 | 5234-5650 | - | Passenger Gene | Antibiotic Resistance |
| tetC | Tn10 | 5738-6331 | + | Passenger Gene | Antibiotic Resistance |
| tet(B) (ARO:3000166) | Tn10 | 6444-7649 | - | Passenger Gene | Antibiotic Resistance |
| tetR (ARO:3003479) | Tn10 | 7728-8354 | + | Passenger Gene | Antibiotic Resistance |
| jemC | Tn10 | 8332-9018 | - | Passenger Gene | Other |
| CPT07_26605 | Tn10 | 9026-9412 | - | Passenger Gene | Hypothetical |
| jemB | Tn10 | 9405-9725 | - | Passenger Gene | Other |
| jemA | Tn10 | 10169-11374 | + | Passenger Gene | Other |
| tnp | IS10L | 11740-12948 | - | Transposase | |
| tnpA | Tn5393.8 | 13000-15828 | + | Transposase | None |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | APH(6)-Id (ARO:3002660) | APH(6)-Id | Tn5393.8 | 281 | 107-952 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | APH(6) (ARO:3000151) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | strB, orfI || strict match to reference sequence for ARO:3002660 (bitscore: 568) | ||||||||||
| Protein Sequence: | MGLMFMPPVF PAHWHVSQPV LIADTFSSLV WKVSLPDGTP AIVKGLKPIE DIADELRGAD YLVWRNGRGA VRLLGRENNL MLLEYAGERM LSHIVAEHGD YQATEIAAEL MAKLYAASEE PLPSALLPIR DRFAALFQRA RDDQNAGCQT DYVHAAIIAD QMMSNASELR GLHGDLHHEN IMFSSRGWLV IDPVGLVGEV GFGAANMFYD PADRDDLCLD PRRIAQMADA FSRALDVDPR RLLDQAYAYG CLSAAWNADG EEEQRDLAIA AAIKQVRQTS Y |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | APH(3'')-Ib (ARO:3002639) | APH(3'')-Ib | Tn5393.8 | 267 | 943-1746 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | APH(3'') (ARO:3000127) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | strA, orfH || perfect match to referencesequence for ARO: 3002639 | ||||||||||
| Protein Sequence: | MNRTNIFFGE SHSDWLPVRG GESGDFVFRR GDGHAFAKIA PASRRGELAG ERDRLIWLKG RGVACPEVIN WQEEQEGACL VITAIPGVPA ADLSGADLLK AWPSMGQQLG AVHSLSVDQC PFERRLSRMF GRAVDVVSRN AVNPDFLPDE DKSTPQLDLL ARVERELPVR LDQERTDMVV CHGDPCMPNF MVDPKTLQCT GLIDLGRLGT ADRYADLALM IANAEENWAA PDEAERAFAV LFNVLGIEAP DRERLAFYLR LDPLTWG |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | orfAB | OrfAB | IS1133 | 376 | 1853-2982 | - |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | fusion protein from -1 programmed frameshifting between orfA and orfB | ||||||||||
| Protein Sequence: | MSLKHSDEFK RDAVRIALTS GLTRRQVASD LSIGLSTLGK WIASISDETK IPTQDTDLLR ENERLRKENR ILREEREILK KAAIFFAVQK L*DFSLLRIT VALSHVHAYV V*WA*QIVVY VHGNAVLHHC ASVVILYF*R IYVSSIGCVW GAMVGRV*QK S*KRWACRLG SVGLDV*CAR ITLQLFERVN SNGQRIVIIP STLHRTY*NK TLAQAHPTRN GQAISLMFGP EKDGSILLLS LTCIPVA*LA GQQVID*SRI LH*GH*IWRW LYANHHRVVF NTQTVGANIA LMNIKSYCSN INCCRP*AGK AIVLITPQ*K ASLNH*RLS* FGADTGKQGE ILRLQSSNI* MAFIIHAEDI QHSAGNRRWH LRKKPL |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | orfB | OrfB | IS1133 | 285 | 1853-2710 | - |
|---|
| Class: | Transposase | ||||||||||
| Function: | Nucleic acid binding (GO:0003676) | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | frameshiftrequired| 5'-end of tnp gene | ||||||||||
| Protein Sequence: | MRFQFITDYR GSLSRSRICR LMGVTDRGLR AWKRRPPSLR QRRDLILLAH IREQHRLCLG SYGRPRMTEE LKALGLQVGQ RRVGRLMRQN NITVVRTRKF KRTTDSHHTF NIAPNLLKQD FSASAPNQKW AGDITYVWTR EGWVYLAVIL DLYSRRVIGW ATGDRLKQDL ALRALNMALA LRKPPPGCIQ HTDRGSQYCA HEYQKLLLKH QLLPSMSGKG NCFDNSAVES FFKSLKAELI WRRHWQTRRD IEIAIFEYIN GFYNPRRRHS TLGWKSPVAF EKKAA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | orfA | OrfA | IS1133 | 91 | 2707-2982 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Regulator | ||||||||||
| Function: | transposase activity (GO:0004803) | ||||||||||
| Comment: | orfA, together with orfB, encodes a fusion protein formed by translational frame-shifting that is the transposase forIS1133 | ||||||||||
| Protein Sequence: | MSLKHSDEFK RDAVRIALTS GLTRRQVASD LSIGLSTLGK WIASISDETK IPTQDTDLLR ENERLRKENR ILREEREILK KAAIFFAVQK L |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn5393.8 | 204 | 3050-3661 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Function: | recombinaseactivity (GO:0000150) | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MLIGYARVSK ADGSQSLDLQ HDALRAAGVE RDNIYDDLAS GGRDDRPGLT ACLKSLRDGD VLVVWKLDRL GRSLAHLVNT VKELSDRKIG LRVLTGKGAQ IDTTTASGRM VFGIFATLAE FERDLIRERT MAGLASARAR GRKGGRKFAL TKAQVRLAQA AMAQRDTSVS DLCKELGIER VTLYRYVGPK GELRDHGKHV LGLT |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnp | Tnp | IS10R | 402 | 4016-5224 | + |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | minor sequence variant-99.9% identical to reference sequence for tnp (IS10R) | ||||||||||
| Protein Sequence: | MCELDILHDS LYQFCPELHL KRLNSLTLAC HALLDCKTLT LTELGRNLPT KARTKHNIKR IDRLLGNRHL HKERLAVYRW HASFICSGNT MPIVLVDWSD IREQKRLMVL RASVALHGRS VTLYEKAFPL SEQCSKKAHD QFLADLASIL PSNTTPLIVS DAGFKVPWYK SVEKLGWYWL SRVRGKVQYA DLGAENWKPI SNLHDMSSSH SKTLGYKRLT KSNPISCQIL LYKSRSKGRK NQRSTRTHCH HPSPKIYSAS AKEPWVLATN LPVEIRTPKQ LVNIYSKRMQ IEETFRDLKS PAYGLGLRHS RTSSSERFDI MLLIALMLQL TC*LAGVHAQ KQGWDKHFQA NTVRNRNVLS TVRLGMEVLR HSGYTITRED LLVAATLLAQ NLFTHGYALG KL |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tetD | TetD | Tn10 | 138 | 5234-5650 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Target: | tetracycline antibiotic (ARO:3000050) | ||||||||||
| Comment: | no match in CARD | ||||||||||
| Protein Sequence: | MYIEQHSRYQ NKANNIQLRY DDKQFHTTVI KDVLLWIEHN LDQSLLLDDV ANKAGYTKWY FQRLFKKVTG VTLASYIRAR RLTKAAVELR LTKKTILEIA LKYQFDSQQS FTRRFKYIFK VTPSYYRRNK LWELEAMH |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tetC | TetC | Tn10 | 197 | 5738-6331 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | DNA binding (GO:0003677) | ||||||||||
| Sequence Family: | Bacterial regulatory proteins, tetR family (Pfam:PF00440) | ||||||||||
| Target: | tetracycline antibiotic (ARO:3000050) | ||||||||||
| Comment: | no match inCARD||99.8% identical to TnCentral reference sequence for tetC (Tn10) | ||||||||||
| Protein Sequence: | MENKNHQQEN FKSTYQSLVN SARILFVEKG YQAVSIDEIS GKALVTKGAF YHHFKNKKQL LSACYKQQLI MIDAYITTKT DLTNGWSALE SIFEHYLDYI IDNNKNLIPI QEVMPIIGWN ELEKISLEYI TGKVNAIVSK LIQENQLKAY DDDVLKNLLN GWFMHIAIHA KNLKELADKK GQFIAIYRGF LLSLKDK |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tet(B) (ARO:3000166) | Tet(B) | Tn10 | 401 | 6444-7649 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic efflux (ARO:0010000) | ||||||||||
| Sequence Family: | major facilitator superfamily (MFS) antibiotic efflux pump (ARO:0010002) | ||||||||||
| Transposase Chemistry: | antibiotic efflux (ARO:0010000) | ||||||||||
| Target: | tetracycline antibiotic (ARO:3000050) | ||||||||||
| Comment: | strict match to reference sequence for ARO:3000166 (bitscore:786)||99.9% identical to TnCentral reference sequence for tet(B) (Tn10)||Synonym: tetB | ||||||||||
| Protein Sequence: | MNSSTKIALV ITLLDAMGIG LIMPVLPTLL REFIASEDIA NHFGVLLALY ALMQVIFAPW LGKMSDRFGR RPVLLLSLIG ASLDYLLLAF SSALWMLYLG RLLSGITGAT GAVAASVIAD TTSASQRVKW FGWLGASFGL GLIAGPIIGG FAGEISPHSP FFIAALLNIV AFLVVMFWFR ETKNTRDNTD TEVGVETQSN SVYITLFKTM PILLIIYFSA QLIGQIPATV WVLFTENRFG WNSMMVGFSL AGLGLLHSVF QAFVAGRIAT KWGEKTAVLL GFIADSSAFA FLAFISEGWL VFPVLILLAG GGIALPALQG VMSIQTKSHQ QGALQGLLVS LTNATGVIGP LLFAVIYNHS LPIWDGWIWI IGLAFYCIII LLSMTFMLTP QAQGSKQETS A |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tetR (ARO:3003479) | TetR | Tn10 | 208 | 7728-8354 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic target alteration (ARO:0001001)||antibiotic efflux (ARO:0010000) | ||||||||||
| Sequence Family: | major facilitator superfamily (MFS) antibiotic efflux pump (ARO:0010002) | ||||||||||
| Transposase Chemistry: | repressor of the tetracycline resistance element | ||||||||||
| Target: | tetracycline antibiotic (ARO:3000050) | ||||||||||
| Comment: | perfect match to reference sequence for ARO:3003479 | ||||||||||
| Protein Sequence: | MMSRLDKSKV INSALELLNE VGIEGLTTRK LAQKLGVEQP TLYWHVKNKR ALLDALAIEM LDRHHTHFCP LEGESWQDFL RNNAKSFRCA LLSHRDGAKV HLGTRPTEKQ YETLENQLAF LCQQGFSLEN ALYALSAVGH FTLGCVLEDQ EHQVAKEERE TPTTDSMPPL LRQAIELFDH QGAEPAFLFG LELIICGLEK QLKCESGS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | jemC | JemC | Tn10 | 228 | 8332-9018 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Sequence Family: | HTH ArsR-type DNA-binding domain (InterPro:IPR001845) | ||||||||||
| Comment: | N-terminal 100 amino acids similar to bacterial transcriptional repressors and metal-binding proteins (PMID:10781570) | ||||||||||
| Protein Sequence: | MANLIRKEVT FESSIAAIGA AMSDISRVKI LSALMDGRAW TATELSSVAN ISASTASSHL SKLLDCQLIT VVAQGKHRYF RLAGKDIAEL MESMMGISLN HGVHAKVSTP VHLRKARTCY DHLAGEVAVK IYDSLCQQQW ITENGSMITL SGIQYFHEMG IDVPSKHSRK ICCACLDWSE RRFHLGGYVG AALFSLYESK GWLTRHLGYR EVTITEKGYA AFKTHFHI |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | CPT07_26605 | CPT07_26605 | Tn10 | 128 | 9026-9412 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Hypothetical | ||||||||||
| Comment: | Amino acid-binding protein | ||||||||||
| Protein Sequence: | MFDVHVVLDN QIGQLALLGK TLGNKGIGLE GGGIFTVGDE CHAHFLVEQG KEAKIALEQA GLLVLAIRTP LIRKLKQEKP GELGEIARVL AENNINILVQ YSDHANQLIL ITDNDSMAAS VTLPWAIK |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | jemB | JemB | Tn10 | 106 | 9405-9725 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Sequence Family: | Antibiotic biosynthesis monooxygenase (Pfam:PF03992) | ||||||||||
| Protein Sequence: | MIAVIFEVQI QPDQQTRYLT LAEELRPLLS HVAGFISIER FQSLATEGKM LSLSWWENEY AVLQWKNHVL HAKAQQEGRE SIFDFYKISI AHITREYSFK KDKDNV |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | jemA | JemA | Tn10 | 401 | 10169-11374 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Function: | glutamate:sodium symporter activity (GO:0015501) | ||||||||||
| Comment: | sodium-dependent glutamate permease | ||||||||||
| Protein Sequence: | MILDASYTLL VACIALLIGM FVVKFTPFLQ KNHIPEAVVG GFIVAIVLLI IDKTSGYSFT FDASLQSLLM LTFFSSIGLS SDFSRLIKGG KPLVLLTIAV TILIAIQNTV GMSMAVMMNE SPFIGLIAGS ITLTGGHGNA GAWGPILADK YDVTGAVELA MACATLGLVL GGLVGGPVAR HLLKKVSIPK TTEQERDTIV EAFEQPSVKR KINANNVIET ISMLIICIVV GGYISALFKD TFLQLPTFVW CLFVGIIIRN TLTHVFKHEV FEPTVDVLGS VALSLFLAMA LMSLKFGQLA SMAGPVLIII AVQTVVMVLF ACFVTFKMMG KDYDAVVISA GHCGFGMGAT PTAIANMQTV TKAFGPSHKA FLVVPMVGAF IVDISNSILI KIFIEIGTYF T |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnp | Tnp | IS10L | 402 | 11740-12948 | - |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MCELDILHDS LYQFCPELHL KRLNSLTLAC HALLDCKTLT LTELGRNLPT KARTKHNIKR IDRLLGNRHL HKERLAVYRW HASFICSGNT MPIVLVDWSD IREQKRLMVL RASVALHGRS VTLYEKAFPL SEQCSKKAHD QFLADLASIL PSNTTPLIVS DAGFKVPWYK SVEKLGWYWL SRVRGKVQYA DLGAENWKPI SNLHDMSSSH SKTLGYKRLT KSNPISCQIL LYKSRSKGRK NQRSTRTHCH HPSPKIYSAS AKEPWILATN LPVEIRTPKQ LVNIYSKRMQ IEETFRDLKS PAYGLGLRHS RTSSSERFDI MLLIALMLQL TCWLAGVHAQ KQGWDKHFQA NTVRNRNVLS TVRLGMEVLR HSGYTITRED SLVAATLLTQ NLFTHGYVLG KL |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn5393.8 | 942 | 13000-15828 | + |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase activity (GO:0004803) | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | probably N-terminal truncation by Tn10 insertion | ||||||||||
| Protein Sequence: | MCIYLNLMIL IKIIRGFISI RLRRRAHNRF GFALQLCVLR YPGRVLAPGE LIPAEVIEFI GAQLGLGADD LVDYAAREET RHEHLAELRG LYGFRTFSGR GASELKEWLF REAEMAVSNE DIARRFVAEC RRTRTVLPAT STIERLCAAA LVDAERRIET RIASRLPMSI REQLLALLEE TADDRVTRFV WLRQFEPGSN SSSANRLLDR LEYLQRIDLP EDLLAGVPAH RVTRLRRQGE RYYADGMRDL PEDRRLAILA VCVSEWQAML ADAVVETHDR IVGRLYRASE RICHAKVADE AGVVRDTLKS FAEIGGALVD AQDDGQPLGD VIASGSGWDG LKTLVAMATR LTATMADDPL NHVLDGYHRF RRYAPRMLRL LDLRAAPVAL PLLEAVTALR TGLNDAAMTS FLRPSSKWHR HLRAQRAGDA RLWEIAVLFH LRDAFRSGDV WLTRSRRYGD LKHALVPAQS IAEGGRLAVP LRPEEWLADR QARLDMRLRE LGRAARAGTI PGGSIENGVL HIEKLEAAAP TGAEDLVLDL YKQIPPTRIT DLLLEVDAAT GFTEAFTHLR TGAPCADRIG LMNVILAEGI NLGLRKMADA TNTHTFWELI RIGRWHVEGE AYDRALAMVV EAQAALPMAR FWGMGTSASS DGQFFVATEQ GEAMNLVNAK YGNTPGLKAY SHVSDQYAPF ATQVIPATAS EAPYILDGLL MNDAGRHIRE QFTDTGGFTD HVFAACAILG YRFAPRIRDL PSKRLYAFNP SAAPAHLRAL IGGKVNQAMI ERNWPDILRI AATIAAGTVA PSQILRKLAS YPRQNELATA LREVGRVERT LFMIDWILDA ELQRRAQIGL NKGEAHHALK RAISFHRRGE IRDRSAEGQH YRIAGMNLLA AIIIFWNTMK LGEVVANQKR DGKLLSPDLL AHVSPLGWEH INLTGEYRWP KP |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| IS1133-Z12167 | IS1133 | insertion sequence | 1815-3046 | + | 1232 |
| Tn10-AF162223 | Tn10 | transposon | 3909-13055 | + | 9147 |
| IS10R-AF162223 | IS10R | insertion sequence | 3909-5237 | + | 1329 |
| IS10L-AF162223 | IS10L | insertion sequence | 11727-13055 | + | 1329 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRR | IS1133 | 1815-1841 | + | 27 | TGGACTTGCA CCGGTTGTGT TCCGGTC |
| IRL | IS1133 | 3020-3046 | - | 27 | TGGACTTGCC TCGGTTTCGT TCCGGTC |
| IRL | IS10R | 3909-3925 | + | 17 | CTGATGAATC CCCTAAT |
| IRR | IS10R | 5216-5237 | - | 22 | CTGAGAGATC CCCTCATAAT TT |
| IRR | IS10L | 11727-11748 | + | 22 | CTGAGAGATC CCCTCATAAT TT |
| IRL | IS10L | 13039-13055 | - | 17 | CTGATGAATC CCCTAAT |
1. Johnson TJ, Thorsness JL, Anderson CP, Lynne AM, Foley SL, Han J, Fricke WF, McDermott PF, White DG, Khatri M, Stell AL, Flores C, Singer RS.
Horizontal gene transfer of a ColV plasmid has resulted in a dominant avian clonal type of Salmonella enterica serovar Kentucky.. PLoS One. 2010 Dec 22;5(12):e15524. doi: 10.1371/journal.pone.0015524. PubMed ID:
21203520
.