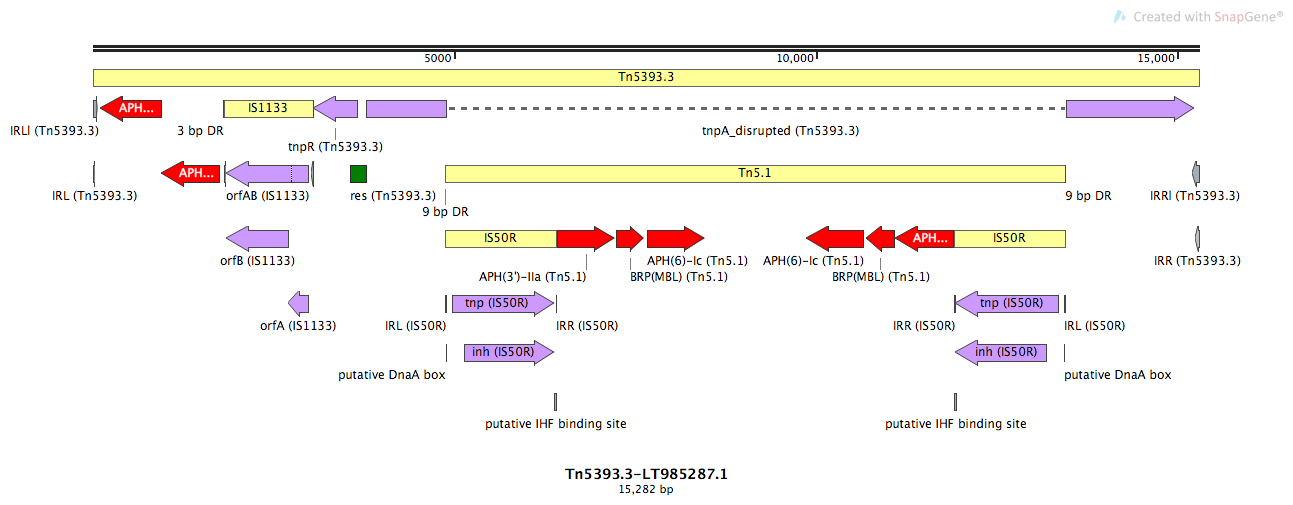
Mobile element type:
Transposon
Name:
Tn5393.3
Synonyms:
Accession:
Tn5393.3-LT985287.1
Family:
Tn3
Group:
Tn163
First isolate:
Yes
Partial:
ND
Evidence of transposition:
No
Host Organism:
Escherichia coli RPC3
Date of Isolation:
2018
Country:
France
Molecular Source:
plasmid RCS69_pI
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRLl | 1-81 | + | 81 | GGGGTCGTTT GCGGGAGAGG GCGAAATCCT ACGCTAAGGC TTTGGCCAAC GATATTCTCC GGTAAGATTG ATGTGTTCCC A |
| IRL | 1-40 | + | 40 | GGGGTCGTTT GCGGGAGAGG GCGAAATCCT ACGCTAAGGC |
| IRRl | 15202-15282 | - | 81 | GGGGTCGTTT GCGGGAGGGG GCGGAATCCT ACGCTAAGGC TTTGGCCAGC GATATTCTCC GGTGAGATTG ATGTGTTCCC A |
| IRR | 15243-15282 | - | 40 | GGGGTCGTTT GCGGGAGGGG GCGGAATCCT ACGCTAAGGC |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res | Tn5393.3 | 3565-3786 | + | 222 | TGTCCCGTTC GACACCTGCG GCGCGCAAGG CGTCGTGCTG CAGGTCGAGA GACTGCGAGC CATCGGCTTT GGAGACGCGG GCATATCCGA TCAGCATGTA TCACAAACGT TGGTTTGAGG CGGCGCTTCG GCCACGATTG CATTGACCTC TGGAAATGTA TCTCAACCAG CTTCATAAAC AAAGCGTCTT GAACGCTATC AGATTTTGAA AAAGGAACAT GT |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| APH(6)-Id (ARO:3002660) | Tn5393.3 | 107-952 | - | Passenger Gene | Antibiotic Resistance |
| orfAB | IS1133 | 1853-2982 | - | Transposase | |
| orfB | IS1133 | 1853-2710 | - | Transposase | |
| orfA | IS1133 | 2707-2982 | - | Accessory Gene | Regulator |
| tnpR | Tn5393.3 | 3050-3661 | - | Accessory Gene | Resolvase |
| tnpA_disrupted | Tn5393.3 | 3787-4908 , 13459-15249 | + | Transposase | |
| tnpA 5'-end | Tn5393.3 | 3787-4908 | + | Transposase | |
| tnp | IS50R | 4972-6402 | + | Transposase | |
| inh | IS50R | 5137-6401 | + | Accessory Gene | Inhibitor |
| APH(3')-IIa (ARO:3002644) | Tn5.1 | 6431-7225 | + | Passenger Gene | Antibiotic Resistance |
| BRP(MBL) (ARO:3001205) | Tn5.1 | 7246-7626 | + | Passenger Gene | Antibiotic Resistance |
| APH(6)-Ic (ARO:3002659) | Tn5.1 | 7665-8465 | + | Passenger Gene | Antibiotic Resistance |
| APH(6)-Ic (ARO:3002659) | Tn5.1 | 9862-10662 | - | Passenger Gene | Antibiotic Resistance |
| BRP(MBL) (ARO:3001205) | Tn5.1 | 10701-11081 | - | Passenger Gene | Antibiotic Resistance |
| APH(3')-IIa (ARO:3002644) | Tn5.1 | 11102-11896 | - | Passenger Gene | Antibiotic Resistance |
| tnp | IS50R | 11925-13355 | - | Transposase | |
| inh | IS50R | 11926-13190 | - | Accessory Gene | Inhibitor |
| tnpA 3'-end | Tn5393.3 | 13459-15249 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | APH(6)-Id (ARO:3002660) | APH(6)-Id | Tn5393.3 | 281 | 107-952 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | APH(6) (ARO:3000151) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | strB, orfI || strict match to reference sequence for ARO:3002660 (bitscore: 568) | ||||||||||
| Protein Sequence: | MGLMFMPPVF PAHWHVSQPV LIADTFSSLV WKVSLPDGTP AIVKGLKPIE DIADELRGAD YLVWRNGRGA VRLLGRENNL MLLEYAGERM LSHIVAEHGD YQATEIAAEL MAKLYAASEE PLPSALLPIR DRFAALFQRA RDDQNAGCQT DYVHAAIIAD QMMSNASELR GLHGDLHHEN IMFSSRGWLV IDPVGLVGEV GFGAANMFYD PADRDDLCLD PRRIAQMADA FSRALDVDPR RLLDQAYAYG CLSAAWNADG EEEQRDLAIA AAIKQVRQTS Y |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | orfAB | OrfAB | IS1133 | 376 | 1853-2982 | - |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | fusion protein from -1 programmed frameshifting between orfA and orfB | ||||||||||
| Protein Sequence: | MSLKHSDEFK RDAVRIALTS GLTRRQVASD LSIGLSTLGK WIASISDETK IPTQDTDLLR ENERLRKENR ILREEREILK KAAIFFAVQK L*DFSLLRIT VALSHVHAYV V*WA*QIVVY VHGNAVLHHC ASVVILYF*R IYVSSIGCVW GAMVGRV*QK S*KRWACRLG SVGLDV*CAR ITLQLFERVN SNGQRIVIIP STLHRTY*NK TLAQAHPTRN GQAISLMFGP EKDGSILLLS LTCIPVA*LA GQQVID*SRI LH*GH*IWRW LYANHHRVVF NTQTVGANIA LMNIKSYCSN INCCRP*AGK AIVLITPQ*K ASLNH*RLS* FGADTGKQGE ILRLQSSNI* MAFIIHAEDI QHSAGNRRWH LRKKPL |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | orfB | OrfB | IS1133 | 285 | 1853-2710 | - |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase activity (GO:0004803) | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | orfB, together with orfA, encodes a fusion protein formed by translational frame-shifting that is the transposase for IS1133 | ||||||||||
| Protein Sequence: | MRFQFITDYR GSLSRSRICR LMGVTDRGLR AWKRRPPSLR QRRDLILLAH IREQHRLCLG SYGRPRMTEE LKALGLQVGQ RRVGRLMRQN NITVVRTRKF KRTTDSHHTF NIAPNLLKQD FSASAPNQKW AGDITYVWTR EGWVYLAVIL DLYSRRVIGW ATGDRLKQDL ALRALNMALA LRKPPPGCIQ HTDRGSQYCA HEYQKLLLKH QLLPSMSGKG NCFDNSAVES FFKSLKAELI WRRHWQTRRD IEIAIFEYIN GFYNPRRRHS TLGWKSPVAF EKKAA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | orfA | OrfA | IS1133 | 91 | 2707-2982 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Regulator | ||||||||||
| Function: | transposase activity (GO:0004803) | ||||||||||
| Comment: | orfA, together with orfB, encodes a fusion protein formed by translational frame-shifting that is the transposase forIS1133 | ||||||||||
| Protein Sequence: | MSLKHSDEFK RDAVRIALTS GLTRRQVASD LSIGLSTLGK WIASISDETK IPTQDTDLLR ENERLRKENR ILREEREILK KAAIFFAVQK L |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn5393.3 | 204 | 3050-3661 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Function: | recombinaseactivity (GO:0000150) | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MLIGYARVSK ADGSQSLDLQ HDALRAAGVE RDNIYDDLAS GGRDDRPGLT ACLKSLRDGD VLVVWKLDRL GRSLAHLVNT VKELSDRKIG LRVLTGKGAQ IDTTTASGRM VFGIFATLAE FERDLIRERT MAGLASARAR GRKGGRKFAL TKAQVRLAQA AMAQRDTSVS DLCKELGIER VTLYRYVGPK GELRDHGKHV LGLT |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA_disrupted | TnpA_disrupted | Tn5393.3 | 970 | 3787-4908 , 13459-15249 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MPRRVTLTDR QKDALLRLPT SQTDLLKHYT LSDEDLGHIR LRRRAHNRFG FALQLCVLRY PGRVLAPGEL IPAEVIEFIG AQLGLGADDL VDYAAREETR HEHLAELRGL YGFRTFSGRG ASELKEWLFR EAEMAVSNED IARRFVAECR RTRTVLPATS TIERLCAAAL VDAERRIETR IASRLPMSIR EQLLALLEET ADDRVTRFVW LRQFEPGSNS SSANRLLDRL EYLQRIDLPE DLLAGVPAHR VTRLRRQGER YYADGMRDLP EDRRLAILAV CVSEWQAMLA DAVVETHDRI VGRLYRASER ICHAKVADEA GVVRDTLKSF AEIGGALVDA QDDGQPLGDV IASGSGWDGL KTLVADSYTQ VAS*MATRLT ATMADDPLNH VLDGYHRFRR YAPRMLRLLD LRAAPVALPL LEAVTALRTG LNDAAMTSFL RPSSKWHRHL RAQRAGDARL WEIAVLFHLR DAFRSGDVWL TRSRRYGDLK HALVPAQSIA EGGRLAVPLR PEEWLADRQA RLDMRLRELG RAARAGTIPG GSIENGVLHI EKLEAAAPTG AEDLVLDLYK QIPPTRITDL LLEVDAATGF TEAFTHLRTG APCADRIGLM NVILAEGINL GLRKMADATN THTFWELIRI GRWHVEGEAY DRALAMVVEA QAALPMARFW GMGTSASSDG QFFVATEQGE AMNLVNAKYG NTPGLKAYSH VSDQYAPFAT QVIPATASEA AYILDGLLMN DAGRHIREQF TDTGGFTDHV FAACAILGYR FAPRIRDLPS KRLYAFNPSA APAHLRALIG GKVNQAMIER NWPDILRIAA TIAAGTVAPS QILRKLASYP RQNELATALR EVGRVERTLF MIDWILDAEL QRRAQIGLNK GEAHHALKRA ISFHRRGEIR DRSAEGQHYR IAGMNLLAAI IIFWNTMKLG EVVANQKRDG KLLSPDLLAH VSPLGWEHIN LTGEYRWPKP |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA 5'-end | N/A | Tn5393.3 | 373 | 3787-4908 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | disruped by insertion of Tn5.1 | ||||||||||
| Protein Sequence: | MPRRVTLTDR QKDALLRLPT SQTDLLKHYT LSDEDLGHIR LRRRAHNRFG FALQLCVLRY PGRVLAPGEL IPAEVIEFIG AQLGLGADDL VDYAAREETR HEHLAELRGL YGFRTFSGRG ASELKEWLFR EAEMAVSNED IARRFVAECR RTRTVLPATS TIERLCAAAL VDAERRIETR IASRLPMSIR EQLLALLEET ADDRVTRFVW LRQFEPGSNS SSANRLLDRL EYLQRIDLPE DLLAGVPAHR VTRLRRQGER YYADGMRDLP EDRRLAILAV CVSEWQAMLA DAVVETHDRI VGRLYRASER ICHAKVADEA GVVRDTLKSF AEIGGALVDA QDDGQPLGDV IASGSGWDGL KTLVADSYTQ VAS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnp | Tnp | IS50R | 476 | 4972-6402 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | 99% identical to tnp (Tn5) | ||||||||||
| Protein Sequence: | MITSALHRAA DWAKSVFSSA ALGDPRRTAR LVNVAAQLAK YSGKSITISS EGSEAMQEGA YRFIRNPNVS AEAIRKAGAM QTVKLAQEFP ELLAIEDTTS LSYRHQVAEE LGKLGSIQDK SRGWWVHSVL LLEATTFRTV GLLHQEWWMR PDDPADADEK ESGKWLAAAA TSRLRMGSMM SNVIAVCDRE ADIHAYLQDK LAHNERFVVR SKHPRKDVES GLYLYDHLKN QPELGGYQIS IPQKGVVDKR GKRKNRPARK ASLSLRSGRI TLKQGNITLN AVLAEEINPP KGETPLKWLL LTSEPVESLA QALRVIDIYT HRWRIEEFHK AWKTGAGAER QRMEEPDNLE RMVSILSFVA VRLLQLRESF TLPQALRAQG LLKEAEHVES QSAETVLTPD ECQLLGYLDK GKRKRKEKAG SLQWAYMAIA RLGGFMDSKR TGIASWGALW EGWEALQSKL DGFLAAKDLM AQGIKI |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | inh | Inh | IS50R | 421 | 5137-6401 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Inhibitor | ||||||||||
| Function: | Inhibitor of transposition | ||||||||||
| Comment: | 99% identical to inh (IS50R)|this is an internal start in the transposase gene from an internal promoter | ||||||||||
| Protein Sequence: | MQEGAYRFIR NPNVSAEAIR KAGAMQTVKL AQEFPELLAI EDTTSLSYRH QVAEELGKLG SIQDKSRGWW VHSVLLLEAT TFRTVGLLHQ EWWMRPDDPA DADEKESGKW LAAAATSRLR MGSMMSNVIA VCDREADIHA YLQDKLAHNE RFVVRSKHPR KDVESGLYLY DHLKNQPELG GYQISIPQKG VVDKRGKRKN RPARKASLSL RSGRITLKQG NITLNAVLAE EINPPKGETP LKWLLLTSEP VESLAQALRV IDIYTHRWRI EEFHKAWKTG AGAERQRMEE PDNLERMVSI LSFVAVRLLQ LRESFTLPQA LRAQGLLKEA EHVESQSAET VLTPDECQLL GYLDKGKRKR KEKAGSLQWA YMAIARLGGF MDSKRTGIAS WGALWEGWEA LQSKLDGFLA AKDLMAQGIK I |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | APH(3')-IIa (ARO:3002644) | APH(3')-IIa | Tn5.1 | 264 | 6431-7225 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | APH(3') (ARO:3000126) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Protein Sequence: | MIEQDGLHAG SPAAWVERLF GYDWAQQTIG CSDAAVFRLS AQGRPVLFVK TDLSGALNEL QDEAARLSWL ATTGVPCAAV LDVVTEAGRD WLLLGEVPGQ DLLSSHLAPA EKVSIMADAM RRLHTLDPAT CPFDHQAKHR IERARTRMEA GLVDQDDLDE EHQGLAPAEL FARLKARMPD GEDLVVTHGD ACLPNIMVEN GRFSGFIDCG RLGVADRYQD IALATRDIAE ELGGEWADRF LVLYGIAAPD SQRIAFYRLL DEFF |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | BRP(MBL) (ARO:3001205) | BRP(MBL) | Tn5.1 | 126 | 7246-7626 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | Bleomycin resistant protein (ARO:3004256) | ||||||||||
| Target: | glycopeptide antibiotic (ARO:3000081) | ||||||||||
| Comment: | loose match 57% to reference sequence for ARO:3001205 (bitscore: 142)||Synonyms: blmS, ble, BRP, blmA, BLMT | ||||||||||
| Protein Sequence: | MTDQATPNLP SRDFDSTAAF YERLGFGIVF RDAGWMILQR GDLMLEFFAH PGLDPLASWF SCCLRLDDLA EFYRQCKSVG IQETSSGYPR IHAPELQEWG GTMAALVDPD GTLLRLIQNE LLAGIS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | APH(6)-Ic (ARO:3002659) | APH(6)-Ic | Tn5.1 | 266 | 7665-8465 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | APH(6) (ARO:3000151) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | perfect match to ARO:3002659 reference sequence||Synonym: str | ||||||||||
| Protein Sequence: | MERWRLLRDG ELLTTHSSWI LPVRQGDMPA MLKVARIPDE EAGYRLLTWW DGQGAARVFA SAAGALLMER ASGAGDLAQI AWSGQDDEAC RILCDTAARL HAPRSGPPPD LHPLQEWFQP LFRLAAEHAA LAPAASVARQ LLAAPREVCP LHGDLHHENV LDFGDRGWLA IDPHGLLGER TFDYANIFTN PDLSDPGRPL AILPGRLEAR LSIVVATTGF EPERLLRWII AWTGLSAAWF IGDGDGEGEG AAIDLAVNAM ARRLLD |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | APH(6)-Ic (ARO:3002659) | APH(6)-Ic | Tn5.1 | 266 | 9862-10662 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | APH(6) (ARO:3000151) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | perfect match to ARO:3002659 reference sequence||Synonym: str | ||||||||||
| Protein Sequence: | MERWRLLRDG ELLTTHSSWI LPVRQGDMPA MLKVARIPDE EAGYRLLTWW DGQGAARVFA SAAGALLMER ASGAGDLAQI AWSGQDDEAC RILCDTAARL HAPRSGPPPD LHPLQEWFQP LFRLAAEHAA LAPAASVARQ LLAAPREVCP LHGDLHHENV LDFGDRGWLA IDPHGLLGER TFDYANIFTN PDLSDPGRPL AILPGRLEAR LSIVVATTGF EPERLLRWII AWTGLSAAWF IGDGDGEGEG AAIDLAVNAM ARRLLD |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | BRP(MBL) (ARO:3001205) | BRP(MBL) | Tn5.1 | 126 | 10701-11081 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | Bleomycin resistant protein (ARO:3004256) | ||||||||||
| Target: | glycopeptide antibiotic (ARO:3000081) | ||||||||||
| Comment: | loose match 57% to reference sequence for ARO:3001205 (bitscore: 142)||Synonyms: blmS, ble, BRP, blmA, BLMT | ||||||||||
| Protein Sequence: | MTDQATPNLP SRDFDSTAAF YERLGFGIVF RDAGWMILQR GDLMLEFFAH PGLDPLASWF SCCLRLDDLA EFYRQCKSVG IQETSSGYPR IHAPELQEWG GTMAALVDPD GTLLRLIQNE LLAGIS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | APH(3')-IIa (ARO:3002644) | APH(3')-IIa | Tn5.1 | 264 | 11102-11896 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | APH(3') (ARO:3000126) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Protein Sequence: | MIEQDGLHAG SPAAWVERLF GYDWAQQTIG CSDAAVFRLS AQGRPVLFVK TDLSGALNEL QDEAARLSWL ATTGVPCAAV LDVVTEAGRD WLLLGEVPGQ DLLSSHLAPA EKVSIMADAM RRLHTLDPAT CPFDHQAKHR IERARTRMEA GLVDQDDLDE EHQGLAPAEL FARLKARMPD GEDLVVTHGD ACLPNIMVEN GRFSGFIDCG RLGVADRYQD IALATRDIAE ELGGEWADRF LVLYGIAAPD SQRIAFYRLL DEFF |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnp | Tnp | IS50R | 476 | 11925-13355 | - |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | 99% identical to tnp (Tn5) | ||||||||||
| Protein Sequence: | MITSALHRAA DWAKSVFSSA ALGDPRRTAR LVNVAAQLAK YSGKSITISS EGSEAMQEGA YRFIRNPNVS AEAIRKAGAM QTVKLAQEFP ELLAIEDTTS LSYRHQVAEE LGKLGSIQDK SRGWWVHSVL LLEATTFRTV GLLHQEWWMR PDDPADADEK ESGKWLAAAA TSRLRMGSMM SNVIAVCDRE ADIHAYLQDK LAHNERFVVR SKHPRKDVES GLYLYDHLKN QPELGGYQIS IPQKGVVDKR GKRKNRPARK ASLSLRSGRI TLKQGNITLN AVLAEEINPP KGETPLKWLL LTSEPVESLA QALRVIDIYT HRWRIEEFHK AWKTGAGAER QRMEEPDNLE RMVSILSFVA VRLLQLRESF TLPQALRAQG LLKEAEHVES QSAETVLTPD ECQLLGYLDK GKRKRKEKAG SLQWAYMAIA RLGGFMDSKR TGIASWGALW EGWEALQSKL DGFLAAKDLM AQGIKI |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | inh | Inh | IS50R | 421 | 11926-13190 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Inhibitor | ||||||||||
| Function: | Inhibitor of transposition | ||||||||||
| Comment: | 99% identical to inh (IS50R)|this is an internal start in the transposase gene from an internal promoter | ||||||||||
| Protein Sequence: | MQEGAYRFIR NPNVSAEAIR KAGAMQTVKL AQEFPELLAI EDTTSLSYRH QVAEELGKLG SIQDKSRGWW VHSVLLLEAT TFRTVGLLHQ EWWMRPDDPA DADEKESGKW LAAAATSRLR MGSMMSNVIA VCDREADIHA YLQDKLAHNE RFVVRSKHPR KDVESGLYLY DHLKNQPELG GYQISIPQKG VVDKRGKRKN RPARKASLSL RSGRITLKQG NITLNAVLAE EINPPKGETP LKWLLLTSEP VESLAQALRV IDIYTHRWRI EEFHKAWKTG AGAERQRMEE PDNLERMVSI LSFVAVRLLQ LRESFTLPQA LRAQGLLKEA EHVESQSAET VLTPDECQLL GYLDKGKRKR KEKAGSLQWA YMAIARLGGF MDSKRTGIAS WGALWEGWEA LQSKLDGFLA AKDLMAQGIK I |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA 3'-end | N/A | Tn5393.3 | 596 | 13459-15249 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | disrupted by insertion of Tn5.1 | ||||||||||
| Protein Sequence: | MATRLTATMA DDPLNHVLDG YHRFRRYAPR MLRLLDLRAA PVALPLLEAV TALRTGLNDA AMTSFLRPSS KWHRHLRAQR AGDARLWEIA VLFHLRDAFR SGDVWLTRSR RYGDLKHALV PAQSIAEGGR LAVPLRPEEW LADRQARLDM RLRELGRAAR AGTIPGGSIE NGVLHIEKLE AAAPTGAEDL VLDLYKQIPP TRITDLLLEV DAATGFTEAF THLRTGAPCA DRIGLMNVIL AEGINLGLRK MADATNTHTF WELIRIGRWH VEGEAYDRAL AMVVEAQAAL PMARFWGMGT SASSDGQFFV ATEQGEAMNL VNAKYGNTPG LKAYSHVSDQ YAPFATQVIP ATASEAAYIL DGLLMNDAGR HIREQFTDTG GFTDHVFAAC AILGYRFAPR IRDLPSKRLY AFNPSAAPAH LRALIGGKVN QAMIERNWPD ILRIAATIAA GTVAPSQILR KLASYPRQNE LATALREVGR VERTLFMIDW ILDAELQRRA QIGLNKGEAH HALKRAISFH RRGEIRDRSA EGQHYRIAGM NLLAAIIIFW NTMKLGEVVA NQKRDGKLLS PDLLAHVSPL GWEHINLTGE YRWPKP |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| IS1133-Z12167 | IS1133 | insertion sequence | 1815-3046 | + | 1232 |
| Tn5.1-LT985287.1 | Tn5.1 | transposon | 4880-13447 | + | 8568 |
| IS50R-U00004.1 | IS50R | insertion sequence | 4880-6413 | + | 1534 |
| IS50R-U00004.1 | IS50R | insertion sequence | 11914-13447 | + | 1534 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRR | IS1133 | 1815-1841 | + | 27 | TGGACTTGCA CCGGTTGTGT TCCGGTC |
| IRL | IS1133 | 3020-3046 | - | 27 | TGGACTTGCC TCGGTTTCGT TCCGGTC |
| IRL | IS50R | 4880-4888 | + | 9 | CTGACTCTT |
| IRR | IS50R | 6405-6413 | - | 9 | CTGTCTCTT |
| IRR | IS50R | 11914-11922 | + | 9 | CTGTCTCTT |
| IRL | IS50R | 13439-13447 | - | 9 | CTGACTCTT |
1. Siguier P, Varani A, Perochon J, Chandler M.
Exploring bacterial insertion sequences with ISfinder: objectives, uses, and future developments.. Methods Mol Biol. 2012;859:91-103. doi: 10.1007/978-1-61779-603-6_5. PubMed ID:
22367867
.
2. Heinzel P, Werbitzky O, Distler J, Piepersberg W. A second streptomycin resistance gene from Streptomyces griseus codes for streptomycin-3"-phosphotransferase. Relationships between antibiotic and protein kinases.. Arch Microbiol. 1988;150(2):184-92. doi: 10.1007/BF00425160. PubMed ID: 2844130 .
3. Blot M, Hauer B, Monnet G. The Tn5 bleomycin resistance gene confers improved survival and growth advantage on Escherichia coli.. Mol Gen Genet. 1994 Mar;242(5):595-601. doi: 10.1007/BF00285283. PubMed ID: 7510018 .
4. Yuasa K, Sugiyama M. Bleomycin-induced beta-lactamase overexpression in Escherichia coli carrying a bleomycin-resistance gene from Streptomyces verticillus and its application to screen bleomycin analogues.. FEMS Microbiol Lett. 1995 Oct 1;132(1-2):61-6. doi: 10.1111/j.1574-6968.1995.tb07811.x. PubMed ID: 7590166 .
5. Reznikoff WS. The Tn5 transposon.. Annu Rev Microbiol. 1993;47:945-63. doi: 10.1146/annurev.mi.47.100193.004501. PubMed ID: 7504907 .
6. Steiniger-White M, Rayment I, Reznikoff WS. Structure/function insights into Tn5 transposition.. Curr Opin Struct Biol. 2004 Feb;14(1):50-7. doi: 10.1016/j.sbi.2004.01.008. PubMed ID: 15102449 .
2. Heinzel P, Werbitzky O, Distler J, Piepersberg W. A second streptomycin resistance gene from Streptomyces griseus codes for streptomycin-3"-phosphotransferase. Relationships between antibiotic and protein kinases.. Arch Microbiol. 1988;150(2):184-92. doi: 10.1007/BF00425160. PubMed ID: 2844130 .
3. Blot M, Hauer B, Monnet G. The Tn5 bleomycin resistance gene confers improved survival and growth advantage on Escherichia coli.. Mol Gen Genet. 1994 Mar;242(5):595-601. doi: 10.1007/BF00285283. PubMed ID: 7510018 .
4. Yuasa K, Sugiyama M. Bleomycin-induced beta-lactamase overexpression in Escherichia coli carrying a bleomycin-resistance gene from Streptomyces verticillus and its application to screen bleomycin analogues.. FEMS Microbiol Lett. 1995 Oct 1;132(1-2):61-6. doi: 10.1111/j.1574-6968.1995.tb07811.x. PubMed ID: 7590166 .
5. Reznikoff WS. The Tn5 transposon.. Annu Rev Microbiol. 1993;47:945-63. doi: 10.1146/annurev.mi.47.100193.004501. PubMed ID: 7504907 .
6. Steiniger-White M, Rayment I, Reznikoff WS. Structure/function insights into Tn5 transposition.. Curr Opin Struct Biol. 2004 Feb;14(1):50-7. doi: 10.1016/j.sbi.2004.01.008. PubMed ID: 15102449 .