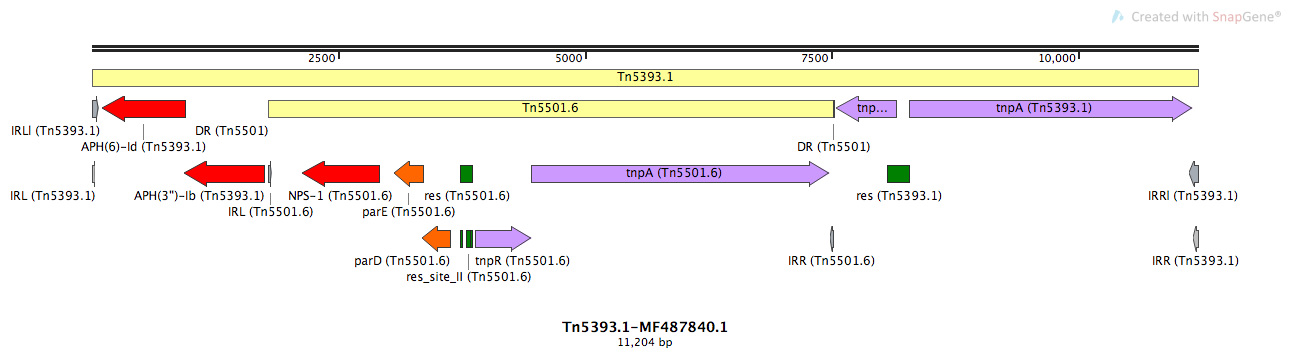
Mobile element type:
Transposon
Name:
Tn5393.1
Synonyms:
Accession:
Tn5393.1-MF487840.1
Family:
Tn3
Group:
Tn163
First isolate:
ND
Partial:
ND
Evidence of transposition:
ND
Host Organism:
Pseudomonas aeruginosa strain PA34 Pseudomonas aeruginosa
Date of Isolation:
2017
Country:
India
Molecular Source:
plasmid PA34
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRLl | 1-81 | + | 81 | GGGGTCGTTT GCGGGAAGGG GCGGAATCCT ACGCTAAGGC TTTGGCCAGC GATATTCTCC GGTGAGATTG ATGTGTTCCC A |
| IRL | 1-40 | + | 40 | GGGGTCGTTT GCGGGAAGGG GCGGAATCCT ACGCTAAGGC |
| IRRl | 11124-11204 | - | 81 | GGGGTCGTTT GCGGGAGGGG GCGGAATCCT ACGCTAAGGC TTTGGCCAGC GATATTCTCC GGTGAGATTG ATGTGTTCCC A |
| IRR | 11165-11204 | - | 40 | GGGGTCGTTT GCGGGAGGGG GCGGAATCCT ACGCTAAGGC |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res | Tn5501.6 | 3733-3863 | + | 131 | GCCTGTCGGA AAACATTTGT TTTGCGACAG GCCTTCAACA GTCCTCTGCA CCAACCTCCG AGTGGCCGCA AAATTGTGCG GAAAACTCTG TCGCCAGACG CTACCATGCG GAAACCCCTT CTTGATGGTT T |
| res_site_I | Tn5501.6 | 3733-3761 | + | 29 | GCCTGTCGGA AAACATTTGT TTTGCGACA |
| res_site_II | Tn5501.6 | 3795-3838 | + | 44 | TGGCCGCAAA ATTGTGCGGA AAACTCTGTC GCCAGACGCT ACCA |
| res_site_III | Tn5501.6 | 3839-3863 | + | 25 | TGCGGAAACC CCTTCTTGAT GGTTT |
| res | Tn5393.1 | 8064-8285 | + | 222 | TGTCCCGTTC GACACCTGCG GCGCGCAAGG TGTCGTGCTG CAGGTCGAGA GACTGCGAGC CATCGGCTTT GGAGACGCGG GCATATCCGA TCAGCATGTA TCACAAACGT TGGTTTGAGG CGGCGCTTCG GCCACGATTG CATTGACCTC TGGAAATGTA TCTCAACCAG CTTCATAAAC AAAGCGTCTT GAACGCTATC AGATTTTGAA AAAGGAACAT GT |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| APH(6)-Id (ARO:3002660) | Tn5393.1 | 107-952 | - | Passenger Gene | Antibiotic Resistance |
| APH(3'')-Ib (ARO:3002639) | Tn5393.1 | 943-1746 | - | Passenger Gene | Antibiotic Resistance |
| NPS-1 (ARO:3004239) | Tn5501.6 | 2136-2918 | - | Passenger Gene | Antibiotic Resistance |
| parE | Tn5501.6 | 3073-3357 | - | Passenger Gene | Toxin |
| parD | Tn5501.6 | 3354-3632 | - | Passenger Gene | Antitoxin |
| tnpR | Tn5501.6 | 3886-4464 | + | Accessory Gene | Resolvase |
| tnpA | Tn5501.6 | 4461-7490 | + | Transposase | |
| tnpR | Tn5393.1 | 7549-8160 | - | Accessory Gene | Resolvase |
| tnpA | Tn5393.1 | 8286-11171 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | APH(6)-Id (ARO:3002660) | APH(6)-Id | Tn5393.1 | 281 | 107-952 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | APH(6) (ARO:3000151) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | strB, orfI || strict match to reference sequence for ARO:3002660 (bitscore: 568) | ||||||||||
| Protein Sequence: | MGLMFMPPVF PAHWHVSQPV LIADTFSSLV WKVSLPDGTP AIVKGLKPIE DIADELRGAD YLVWRNGRGA VRLLGRENNL MLLEYAGERM LSHIVAEHGD YQATEIAAEL MAKLYAASEE PLPSALLPIR DRFAALFQRA RDDQNAGCQT DYVHAAIIAD QMMSNASELR GLHGDLHHEN IMFSSRGWLV IDPVGLVGEV GFGAANMFYD PADRDDLCLD PRRIAQMADA FSRALDVDPR RLLDQAYAYG CLSAAWNADG EEEQRDLAIA AAIKQVRQTS Y |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | APH(3'')-Ib (ARO:3002639) | APH(3'')-Ib | Tn5393.1 | 267 | 943-1746 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | APH(3'') (ARO:3000127) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | strA, orfH || perfect match to referencesequence for ARO: 3002639 | ||||||||||
| Protein Sequence: | MNRTNIFFGE SHSDWLPVRG GESGDFVFRR GDGHAFAKIA PASRRGELAG ERDRLIWLKG RGVACPEVIN WQEEQEGACL VITAIPGVPA ADLSGADLLK AWPSMGQQLG AVHSLSVDQC PFERRLSRMF GRAVDVVSRN AVNPDFLPDE DKSTPQLDLL ARVERELPVR LDQERTDMVV CHGDPCMPNF MVDPKTLQCT GLIDLGRLGT ADRYADLALM IANAEENWAA PDEAERAFAV LFNVLGIEAP DRERLAFYLR LDPLTWG |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | NPS-1 (ARO:3004239) | NPS-1 | Tn5501.6 | 260 | 2136-2918 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | NPS beta-lactamase (ARO:3004240) | ||||||||||
| Target: | cephalosporin (ARO:0000032)||penam (ARO:3000008) | ||||||||||
| Comment: | strict match to reference for ARO:3004240 (bitscore:538)|| | ||||||||||
| Protein Sequence: | MLKSTLLAFG LFIALSARAE NQAIAQLFQR AGVDGTIVIE SLTTRQRLVH NDPRAQQRYP AASTFKVLNT LIALEEGAIS GENQIFHWNG TQYSIANWNQ DQTLDSAFKV SCVWCYQQIA LRVGALKYPA YIQQTNYGHL LEPFNGTEFW LDGSLTISAE EQVAFLRRVV ERKLPFKASS YDSLKKVMFA DENAQYRLYA KTGWATRITP SVGWYVGYVE AQDDVWLFAL NLATRDANDL PLRTQIAKDA LKAIGAFHAK |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | parE | ParE | Tn5501.6 | 94 | 3073-3357 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Toxin | ||||||||||
| Sequence Family: | ParE_toxin (Pfam:PF05016) | ||||||||||
| Target: | DNA gyrase | ||||||||||
| Protein Sequence: | VRVVWTPEAQ QDRADVWDYI AADNPRAAAR MDEIFSDAAA RLIQHPMLGK PGKIPGTREL IPHESYRLVY QIDGETVWIL TLVHTARLWP PVRD |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | parD | ParD | Tn5501.6 | 92 | 3354-3632 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antitoxin | ||||||||||
| Sequence Family: | parD (PDB:4Q2U) | ||||||||||
| Comment: | RelB/ParD/CcdA/DinJ | ||||||||||
| Protein Sequence: | MSKQAVFTMK LEPELRAEFM AEAEAAHRPA SQVLRELMRE FVQRQRESRE YDEFLRRKVE AGRASMRAGL GRSNDEVEAE FAARRASVAS QA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn5501.6 | 192 | 3886-4464 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Function: | recombinaseactivity (GO:0000150) | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MRVSSDSDRQ STNLQRDALL AVGVDARHLF EDHASGAKDD RAGLARALEF VRPGDVLVVW KLDRLGRSLS HLLAIVTSLK EKQVAFRSLT ENLDTTTPSG EFLFQVFGAL AQYERALIQE RVVAGLAAAR KRGRIGGRPQ AITGEKLEAI VAALDGGMSK AAVCRNFGVK RTTLIETLAR VGWTGSRGAS SR |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn5501.6 | 1009 | 4461-7490 | + |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase activity (GO:0004803) | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MTTKSERLTV LSDAEQEALY GLPDFDDAQR LEYLALTETE LALASSRPGL HAQVYCILQI GYFKAKHAFF RFDWSEVEHD CAFVLSRYFH GESFEHKPIS KHEHYTQREW IADLFGYRPW AAEFLPQLAQ QAAQTVRRDV MPGFIAAELI VWLNEHKIIR PGYTTLQELV SEALSAERRR LAGLLSEVLD ESAKAALGRL LVRDDTLSQL AALKQDAKDF GWRQMAHERE KRATLEPLHR IAKALLPKLG VSQQNLLYYA SLANFYTVHD LRNLKADQTY LYLLCYAWVR YRQLSDNLVD AMAYHMKQLE DESSAGAKQS FVAEQVRRQQ DTPQVGRLLS LYIDDSVPDP TPFGDVRQRA YKIMPRDTLQ TTAQRMSVKP VSKLALHWQA VDGLAERIRR HLRPLYVALD LAGTDPGSPW LVALAWAKDV FAKQQRLSQR PLAECPAATL PKRLRPYLLT FDADGKPTGL RADRYEFWLY RQVRKRFQSG ELYLDDSLQH RHFSDELVSL DEKADVLAQI DIPFLRQPLD AQLDALATEL RSQWLAFNRE LKQGKLTHLE YDKDTQKLTW RKPRGENQKA RERAFYEQLP FCDVADVFRF VNGQCQFLSA LTPLQPRYAK KVADADSLMA VIIAQAMNHG NQVMARTSDI PYHVLESAYQ QYLRHATLHA ANDCISNAIA ALPIFPYYSF DLDALYGAVD GQKFGVARPT VKARHSRKYF GRGKGVVAYT LLCNHVPLNG YLIGAHDYEA HHVFDIWYRN TSDIVPTAIT GDMHSVNKAN FAILHWFGLR FEPRFTDLGD QLKELYSADD PALYDQCLIR PAGRIDRDLI VSEKPNLDQI VATLGLKEMT QGTLIRKLCT YTAPNPTRRA VFEFDKLIRS IYTLRYLRDP QLERNVHRSQ NRIESYHQLR STIAQVGGKK ELTGRTDIEI EISNQCARLI ANAVIFYNSA ILSRLLMNYE ASGNAKAHAL LTQISPAAWR HILLNGHYTF QSDGKMIDLD ALVAGLELG |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn5393.1 | 204 | 7549-8160 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Function: | recombinaseactivity (GO:0000150) | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MLIGYARVSK ADGSQSLDLQ HDTLRAAGVE RDNIYDDLAS GGRDDRPGLT ACLKSLRDGD VLVVWKLDRL GRSLAHLVNT VKELSDRKIG LRVLTGKGAQ IDTTTASGRM VFGIFATLAE FERDLIRERT MAGLASARAR GRKGGRKFAL TKAQVRLAQA AMAQRDTSVS DLCKELGIER VTLYRYVGPK GELRDHGKHV LGLT |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn5393.1 | 961 | 8286-11171 | + |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase activity (GO:0004803) | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MPRRVTLTDR QKDALLRLPT SQTDLLKHYT LSDEDLGHIR LRRRAHNRFG FALQLCVLRY PGRVLAPGEL IPAEVIEFIG AQLGLGADDL VDYAAREETR HEHLAELRGL YGFRTFSGRG ASELKEWLFR EAEMAVSNED IARRFVAECR RTRTVLPATS TIERLCAAAL VDAERRIETR IAIRLPMSIR EQLLALLEET ADDRVTRFVW LRQFEPGSNS SSANRLLDRL EYLQRVDLPE DLLAGVPAHR VTRLRRQGER YYADGMRDLP EDRRLAILAV CVSEWQAMLA DAVVETHDRI VGRLYRASER ICHAKVADEA GVVRDTLKSF AEIGGALVDA QDDGQPLGDV IASGSGWDGF KTLVAMATRL TATMADDPLN HVLDGYHRFR RYAPRMLRLL DLRAAPVALP LLEAVTALRT GLNDAAMTSF LRPSSKWHRH LRAQRAGDAR LWEIAVLFHL RDAFRSGDVW LTRSRRYGDL KHALVPAQAI AEGGRLAVPL RPEEWLADRQ ARLDMRLREL GRAARAGTIP GGSIENGVLH IEKLEAAAPT GAEDLVLDLY KQIPPTRITD LLLEVDAATG FTEAFTHLRT GAPCADRIGL MNVILAEGIN LGLRKMADAT NTHTFWELIR IGRWHVEGEA YDRALAMVVE AQAALPMARF WGMGTSASSD GQFFVATEQG EAMNLVNAKY GNTPGLKAYS HVSDQYAPFA TQVIPATASE APYILDGLLM NDAGRHIREQ FTDTGGFTDH VFAACAILGY RFAPRIRDLP SKRLYAFNPS AAPAHLRALI GGKVNQAMIE RNWPDTLRIA ATIAAGTVAP SQILRKLASY PRQNELATAL REVGRVERTL FMIDWILDAE LQRRAQIGLN KGEAHHALKR AISFHRRGEI RDRSAEGQHY RIAGMNLLAA IIIFWNTMKL GEVVANQKRD GKLLSPDLLA HVSPLGWEHI NLTGEYRWPK P |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| Tn5501.6-MF487840.1 | Tn5501.6 | transposon | 1794-7522 | + | 5729 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRL | Tn5501.6 | 1794-1831 | + | 38 | GGGGTTCTAA GCCGGAACCG CCGAAAATTC CGTCAGCC |
| IRR | Tn5501.6 | 7485-7522 | - | 38 | GGGGTTCTAA GCCGGAACCG CCGAAATTTC CGTCATCC |
1. Subedi D, Vijay AK, Kohli GS, Rice SA, Willcox M.
Nucleotide sequence analysis of NPS-1 beta-lactamase and a novel integron (In1427)-carrying transposon in an MDR Pseudomonas aeruginosa keratitis strain.. J Antimicrob Chemother. 2018 Jun 1;73(6):1724-1726. doi: 10.1093/jac/dky073. PubMed ID:
29547943
.