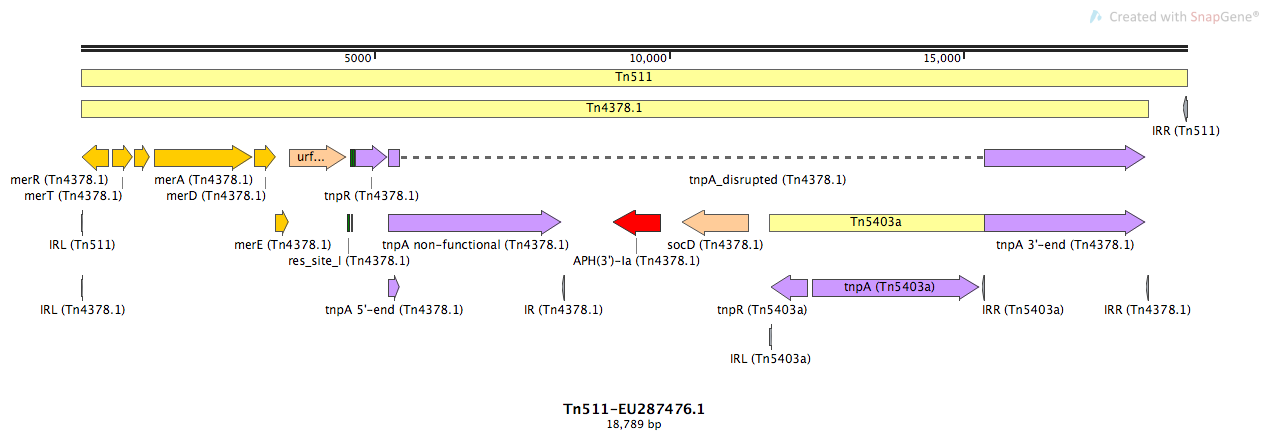
Mobile element type:
Transposon
Name:
Tn511
Synonyms:
Accession:
Tn511-EU287476.1
Family:
Tn3
Group:
Tn163
First isolate:
Yes
Partial:
ND
Evidence of transposition:
ND
Host Organism:
Proteus mirabilis
Date of Isolation:
Country:
Molecular Source:
plasmid R772
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-38 | + | 38 | GGGGTCGTCT CAGAAAACGG AAAATAAAGC ACGCTAAG |
| IRR | 18752-18789 | - | 38 | GGGGTCGCCT CAGAAAACGG AAAATAAAGC ACGCTAAG |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res_site_I | Tn4378.1 | 4527-4557 | + | 31 | CGTCAGATTG AGGCATACCC TAACCGGATG T |
| res_site_II | Tn4378.1 | 4578-4612 | + | 35 | CGTCAGAATA GAGTCGGTTG TGTTATTTAT TGACA |
| res_minus_35 | Tn4378.1 | 4607-4612 | + | 6 | TTGACA |
| res_site_III | Tn4378.1 | 4615-4646 | + | 32 | AGCTGAAAAA GGTCATAGAT TTCTTCCTGA CA |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| merR | Tn4378.1 | 34-468 | - | Passenger Gene | Heavy Metal Resistance |
| merT | Tn4378.1 | 540-890 | + | Passenger Gene | Heavy Metal Resistance |
| merP | Tn4378.1 | 903-1178 | + | Passenger Gene | Heavy Metal Resistance |
| merA | Tn4378.1 | 1250-2935 | + | Passenger Gene | Heavy Metal Resistance |
| merD | Tn4378.1 | 2953-3318 | + | Passenger Gene | Heavy Metal Resistance |
| merE | Tn4378.1 | 3315-3551 | + | Passenger Gene | Heavy Metal Resistance |
| urfM | Tn4378.1 | 3548-4537 | + | Passenger Gene | Other |
| tnpR | Tn4378.1 | 4667-5227 | + | Accessory Gene | Resolvase |
| tnpA non-functional | Tn4378.1 | 5230-8195 | + | Transposase | |
| tnpA 5'-end | Tn4378.1 | 5230-5439 | + | Transposase | |
| APH(3')-Ia (ARO:3002641) | Tn4378.1 | 9047-9862 | - | Passenger Gene | Antibiotic Resistance |
| socD | Tn4378.1 | 10230-11348 | - | Passenger Gene | Other |
| tnpR | Tn5403a | 11740-12345 | - | Accessory Gene | Resolvase |
| tnpA | Tn5403a | 12440-15298 | + | Transposase | |
| tnpA 3'-end | Tn4378.1 | 15370-18119 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merR | MerR | Tn4378.1 | 144 | 34-468 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | activator-repressor of mer operon | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MENNLENLTI GVFAKAAGVN VETIRFYQRK GLLLEPDKPY GSIRRYGEAD VTRVRFVKSA QRLGFSLDEI AELLRLEDGT HCEEASSLAE HKLKDVREKM ADLARMEAVL SELVCACHAR RGNVSCPLIA SLQGGASLAG SAMP |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merT | MerT | Tn4378.1 | 116 | 540-890 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | cytosolic mercuric ion transport protein | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MSEPKTGRGA PFTGGLAAIL ASACCLGPLV LIALGFSGAW IGNLAVLEPY RPIFIGVALV ALFFAWRRIY RQAAACKPGE VCAIPQVRAT YKLIFWIVAA LVLVALGFPY VMPFFY |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merP | MerP | Tn4378.1 | 91 | 903-1178 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercury transport | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MKKLFASLAL AAVVAPVWAA TQTVTLSVPG MTCSACPITV KKAISKVEGV SKVDVTFETR QAVVTFDDAK TSVQKLTKAT ADAGYPSSVK Q |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merA | MerA | Tn4378.1 | 561 | 1250-2935 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercuric ion reductase | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MTHLKITGMT CDSCAAHVKE ALEKVPGVQS ALVSYPKGTA QLAIVPGTSP DALTAAVAGL GYKATLADAP LADNRVGLLD KVRGWMAAAE KHSGNEPPVQ VAVIGSGGAA MAAALKAVEQ GAQVTLIERG TIGGTCVNVG CVPSKIMIRA AHIAHLRRES PFDGGIAATV PTIDRSKLLA QQQARVDELR HAKYEGILGG NPAITVVHGE ARFKDDQSLT VRLNEGGERV VMFDRCLVAT GASPAVPPIP GLKESPYWTS TEALASDTIP ERLAVIGSSV VALELAQAFA RLGSKVTVLA RNTLFFREDP AIGEAVTAAF RAEGIEVLEH TQASQVAHMD GEFVLTTTHG ELRADKLLVA TGRTPNTRSL ALDAAGVTVN AQGAIAIDQG MRTSNPNIYA AGDCTDQPQF VYVAAAAGTR AAINMTGGDA ALDLTAMPAV VFTDPQVATV GYSEAEAHHD GIETDSRTLT LDNVPRALAN FDTRGFIKLV IEEGSHRLIG VQAVAPEAGE LIQTAALAIR NRMTVQELAD QLFPYLTMVE GLKLAAQTFN KDVMQLSCCA G |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merD | MerD | Tn4378.1 | 121 | 2953-3318 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | secondary regulatory protein | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MNAYTVSRLA LDAGVSVHIV RDYLLRGLLR PVACTPGGYG LFDDAALQRL CFVRAAFEAG IGLDALARLC RALDAADGDE AAAQLALLRQ FVERRREALA DLEVQLATLP TEPAQHAESL P |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merE | MerE | Tn4378.1 | 78 | 3315-3551 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercury transport | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MNNPERLPSE THKPITGYLW GGLAVLTCPC HLPILAVVLA GTTAGAFLGE HWVIAALGLT GLFLLSLSRA LRAFRERE |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | urfM | UrfM | Tn4378.1 | 329 | 3548-4537 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Function: | possible diguanylate phosphodiesterase | ||||||||||
| Sequence Family: | EAL (Pfam:PF00563)||DUF3330 (Pfam:PF11809) | ||||||||||
| Comment: | similar to urfM from E.coli | ||||||||||
| Protein Sequence: | MSAFRPDGWT TPELAQAVER GQLELHYQPV VDLRSGGIVG AEALLRWRHP TLGLLPPGQF LPVVESSGLM PEIGAWVLGE ACRQMRDWRM LAWRPFRLAV NVSASQVGPD FDGWVKGVLA DAELPAEYLE IELTESVAFG DPAIFPALDA LRQIGVRFAA DDFGTGYSCL QHLKCCPIST LKIDQSFVAG LANDRRDQTI VHTVIQLAHG LGMDVVAEGV ETSASLDLLR QADCDTGQGF LFAKPMPAAA FAVFVSQWRG ATMNASDSTT TSCCVCCKEI PLDAAFTPEG AEYVEHFCGL ECYQRFEARA KTGNETDADP NACDSLPSD |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn4378.1 | 186 | 4667-5227 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Function: | resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MQGQRIGYVR VSSFDQNPER QLEHVEVGKV FTDKASGKDT QRPELDSLLA FVREGDTVVV HSMDRLARNL DDLRRLVQKL TKRGVRIEFV KESLTFTGED SPMANLMLSV MGAFAEFERA LIRERQREGI ALAKQRGAYR GRKKALSPEQ VADLRQRAAA GEQKAKLARE FGVSRETLYQ YLRADQ |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA non-functional | TnpA non-functional | Tn4378.1 | 988 | 5230-8195 | + |
|---|
| Class: | Transposase | ||||||||||
| Comment: | non-functional transposase due to internal stop codon || appears to be a fusion of the first 70 codons of tnpA (Tn4378) and another unknown tnpA | ||||||||||
| Protein Sequence: | MPRRSILSAA ERESLLALPD TKDDLIRYYT FSDTDLSIIR QRRGPANRLG FAVQLCYLRF PGILLGVDEP DEPPFRPAET VADQLKVSVE SWASTGSGSR PGASIWSSAN GVRLPALHHE PLPAGRPHAD RAGHANRQGH CAGQRLDRAS AAAVGHSACP QRRRAGERRS DHPRQPAHLL RLGRTTVGRA SPPPRRSAQA PGQRQDDLAG LAAPVTRQAQ FAAYAGAHRT TQGMAGARSA YRHRAADPPK PAAQDRPRGR PDDTRRPGQV RGAAALRDPG GARH*RHGHR HRRNHRPARP HPGQAVQRRQ EQASAAVPGV RQGDQRQGAA VRAYRSGTDR GQAIGPRSVC RHRGRHVLGR LRRERHRSAE ARAARGLRFP APHRRELRHA ASLRAGIPRR AQAAGRARCQ GCAGGHRSAA QHEQRQRPQG ARRRANRFHQ AALAEAGDDR HRHRSALLRT VRAVGDEKRP ALRRHLGAGI APVQGLRGLP GATREIRQPQ AGQRIAAGRG HRLRPVPA*P ADAAGNAARH RQPHGAGQRA AGRHHHGVGP EDHAARCGGA RHRAGADRPD SNDPAARQDH RTAAGGRRMD RLHPALRAPE IGRPGQGQEP AADHDPGRRH QPGSDQDGGV LPRNDLRQAR LAPSLAYPRR NLFVGAGRTG QCAVPASLRR ALGRRHHVIV GRPEFPNRQQ GREHWPHQPE IWQQSWADFL HPHLRPVRAI PHQGGSCRRA RLDYVLDGCC TRVRLRIEST TPIRQDSPIM YLADALLGVR LRRASATWAT PSCSSPRATP STTRSSR*LA ATD*TSRLFA PIGMKFYGWP RRSSRAR*RL R*CCASSAAI RARTAWPWPC ASWGVSSARC SSWIGCKAWS CAVACTLG*T RAKPAMRWPA PCSSTVWVKS ATAALSSSAT VPAASTW*RR PSCYGTRSIW NGLRTRCGAT ATPSMTRCCS TCRRSAGSTS T*PAITSGAA APRSARASSG RCGRCNLL |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA 5'-end | N/A | Tn4378.1 | 70 | 5230-5439 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | tnpA ORF interrupted | ||||||||||
| Protein Sequence: | MPRRSILSAA ERESLLALPD TKDDLIRYYT FSDTDLSIIR QRRGPANRLG FAVQLCYLRF PGILLGVDEP |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | APH(3')-Ia (ARO:3002641) | APH(3')-Ia | Tn4378.1 | 271 | 9047-9862 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | APH(3') (ARO:3000126) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | strict match to reference sequence for ARO:3002641 (bitscore: 550)||Synonyms: aphA-1,apha1-1AB, APH(3')-Ic, apha7 | ||||||||||
| Protein Sequence: | MSHIQRETSC SRPRLNSNLD ADLYGYRWAR DNVGQSGATI YRLYGKPDAP ELFLKHGKGS VANDVTDEMV RLNWLTAFMP LPTIKHFIRT PDDAWLLTTA IPGKTAFQAL EEYPDSRENI VDALAAFLRR LHSIPLCNCP FNSDRVFRLA QAQSRMNNGL VDASDFDDER NGCPVEQVWK EMHKLLPFSP DSVVTHGDFS LDNLIFDEGK LIGCIDVGRV GIADRYQDLA ILWNCLGEFS PSLQKRLFHK YGIDNPDMNK LQFHLMLDEF F |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | socD | SocD | Tn4378.1 | 372 | 10230-11348 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Comment: | putative santhopine-degrading protein ||FAD-binding oxidoreductase | ||||||||||
| Protein Sequence: | MSDKTVAKTV IVLGGGILGV STAWQLAQAG AQVTLVTEAA LCSGATGRSL SWLNSAGERS QPYHALRIAG IDRYRTLFAR HPQLDWLRFD GAIYWAADDD AGTKARHHYE KAQGYDSKLI NRATVGDVDA QVNPASLGHV AIANPGEGWV SLPHLVDHLV QAFRALGGEI IENTGKASVI TKDGRACGIR SEKHGALLAD QVLVACGPWT PEVVAPLGVQ IPNGSPVSML VTSQPCDLAP QVVLNTPRAA VRPNPGNTIA VDHDWYEEHI VAQGDGQYRI DTSVINQLMA EAGNLIGDGA PLTANSCKIG LKPIPGDGEP VVGELQKVPG CFVVFTHSGA TLALILGEML TQEMLTGVKH PMLATFRPER FS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn5403a | 201 | 11740-12345 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MGHRAAIYCR VSTADQSCER QEFDLRAFAG RAGYDVVGIF KETGSGTKLD RAERKKVLAL AQSRQIDAIL VTELSRWGRS TLDLLNTLRE LENWKVSVIA MNGMAFDLSS PYGRMLATFL SGIAEFERDL ISERVKSGLA VAKARGKRLG RQAGVRPKSD RLLPKVVAMR AEGRSYRWIA RELGISKNTV ADIVQRHRAN A |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn5403a | 952 | 12440-15298 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MSRRHIFTER QRAALFDLPT DELSLLKFYT LGDDDLENIR QRRRPENRIG FALQLCALRY PGRALAPGEM IPREILSFVG AQLGVPADAL LTYATRRQTR QQHMDTLREI YGYKTFTGRG ARDLRKWTFG QAEDARSNED LAHRFIVRCR ETSTILPAVS TIERLCADAL VAAERRIETR IVENLTADVR DHLDKLLSEM LAGNISRFIW LRNFEVGNNS AAANRLLDRL EFLRTLNINH SALASIPAHR IARLRRQGER YFTDGLRDIT SDRRWAILAV CVVEWEAAIA DAIVETHDRI VGKTWREAKR QHDETISGSK ATLADTIRTF TALGASLLEA RSDGTPLEMA VASSVAWDRL AQLVATGTQL SNTLADEPLA YVGQGYHRFR RYAPRMLRCL KLEAAPVAGP LVAAALSIGE MKGVASPERR FLRPSSKWNR HLRAQEKGDT RLWEVAVLFH LRDAFRSGDV WLAHSRRYGD LKQVLVPMIA AQENAKLAVP SNPQDWLADR KARLTIALKR LARAARNGTI PHGSIEDGTL RIDRLTADVP DGAEALILDL YRRMPSVRIT DMLLEVDAAL GFTDAFTHLR TGAPCRDRIG LLNVLLAEGL NLGLRKMAEA TNTHDYWQLS RLARWHVESE AMNQALAIVV AAQGKLPMSR VWGMGTSASS DGQFFPTARH GEAMNMVNAK YGSVPGLKAY THVSDQFAPF ACQSIPATVS EAPYILDGLL MNEVGRHVRE QYADTAGFTD HLFGASSLLG YNLVLRIRDL PSKRLYVFNP DTTPRELRKL VGGKAREDLI VANWPDIFRC AATMTAGKIR PSQLLRKLAS YPRQNNLAVA LREVGRIERT LFIIEWILDT DMQRRAQIGL NKGEAHHALK NALRIGRQGE IRDRTTEGQH YRIAGLNLLT AVIIYWNTVH LGHAVTERRN EGLDVPPEFL PHISPLAGRT FY |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA 3'-end | N/A | Tn4378.1 | 916 | 15370-18119 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | tnpA ORF interrupted | ||||||||||
| Protein Sequence: | PLLKLVADQL KVSVESWAST GSGSRPGASI WSSANGVRLP ALHHEPLPAG RPHADRAGHA NRQGHCAGQR LDRASAAAVG HSACPQRRRA GERRSDHPRQ PAHLLRLGRT TVGRASPPPR RSAQAPGQRQ DDLAGLAAPV TRQAQFAAYA GAHRTTQGMA GARSAYRHRA ADPPKPAAQD RPRGRPDDTR RPGQVRGAAA LRDPGGARH* RHGHRHRRNH RPARPHPGQA VQRRQEQASA AVPGVRQGDQ RQGAAVRAYR SGTDRGQAIG PRSVCRHRGR HVLGRLRRER HRSAEARAAR GLRFPAPHRR ELRHAASLRA GIPRRAQAAG RARCQGCAGG HRSAAQHEQR QRPQGARRRA NRFHQAALAE AGDDRHRHRS ALLRTVRAVG NEKRPALRRH LGAGIAPVQG LRGLPGATRE IRQPQAGQRI AAGRGHRLRP VPA*PADAAG NAARHRQPHG AGQRAAGRHH HGVGPEDHAA RCGGARHRAG ADRPDSNDPA ARQDHRTAAG GRRMDRLHPA LRAPEIGRPG QGQEPAADHD PGRRHQPGSD QDGGVLPRND LRQARLAPSL AYPRRNLFVG AGRTGQCAVP ASLRRALGRR HHVIVGRPEF PNRQQGREHW PHQPEIWQQS WADFLHPHLR PVRAIPHQGG QCRRARLDLC ARRLAVPRVR PAHRGALHRY GRIHRSCIWP DAPAGLPLCA AHPRPGRHQA VHPQGRHRLR RAQADD*QRQ TEHQGYSRPL G*NSTAGHVD QAGHGDGFAD AAQARQLSAP ERPGRGPARA GAYRAHAVHP GLVAKRGAAP SRARWAEQGR SPQCAGPRRV LQPSG*NPRP QL*AAALPCQ RPQPGDGGRR AMEHGLSGTG CARAAGQRPR RR*RAVAVPV AARLGAHQPD RRLPLAQQRQ DRRGQVQAAA AVATCL |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| Tn4378.1-EU287476.1 | Tn4378.1 | transposon | 1-18151 | + | 18151 |
| Tn5403a-EU287476.1 | Tn5403a | transposon | 11707-15369 | + | 3663 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRL | Tn4378.1 | 1-38 | + | 38 | GGGGTCGTCT CAGAAAACGG AAAATAAAGC ACGCTAAG |
| IR | Tn4378.1 | 8191-8228 | - | 38 | GGGGTCGTCT CAGAAAACGG AAAATAAAGC ACGCTAAG |
| IRL | Tn5403a | 11707-11752 | + | 46 | GGGGTCGGTT CCGGCTGAGG GCGAAATGAC ACCCTAAGCG TTAGCT |
| IRR | Tn5403a | 15324-15369 | - | 46 | GGGGTCGGTT CCGGCTGAGG GCGAAATGAC ACCCTAAGCT TTCGGT |
| IRR | Tn4378.1 | 18114-18151 | - | 38 | GGGGTCGTCT CAGAAAACGG AAAATAAAGC ACGCTAAG |
1. Petrovski S, Stanisich VA.
Embedded elements in the IncPbeta plasmids R772 and R906 can be mobilized and can serve as a source of diverse and novel elements.. Microbiology (Reading). 2011 Jun;157(Pt 6):1714-1725. doi: 10.1099/mic.0.047761-0. Epub 2011 Mar 10. PubMed ID:
21393370
.
2. Coetzee JN. Mobilization of the Proteus mirabilis chromosome by R plasmid R772.. J Gen Microbiol. 1978 Sep;108(1):103-9. doi: 10.1099/00221287-108-1-103. PubMed ID: 357678 .
2. Coetzee JN. Mobilization of the Proteus mirabilis chromosome by R plasmid R772.. J Gen Microbiol. 1978 Sep;108(1):103-9. doi: 10.1099/00221287-108-1-103. PubMed ID: 357678 .