Mobile element type:
Transposon
Name:
Tn5041
Synonyms:
Accession:
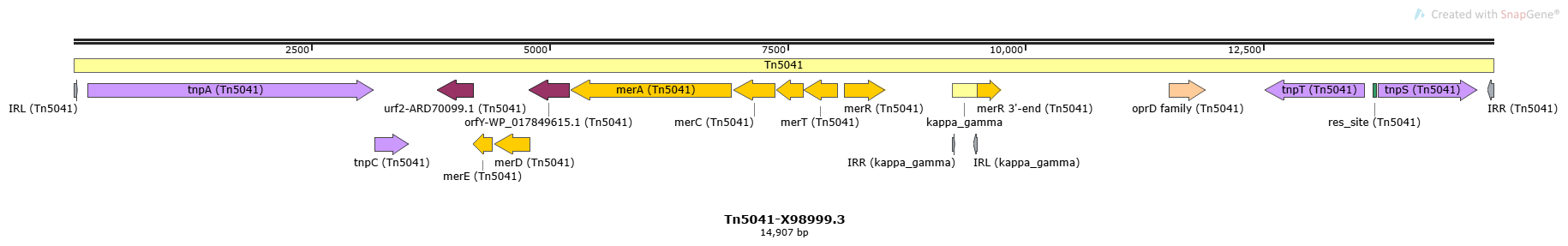
Tn5041-X98999.3
Family:
Tn3
Group:
Tn4651
First isolate:
Yes
Partial:
ND
Evidence of transposition:
Yes
Host Organism:
Pseudomonas sp.
Date of Isolation:
1997
Country:
Central asia
Molecular Source:
plasmid
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-47 | + | 47 | GGGGTTATGC CGAGATAAGG CAAAAATTAG GACATTCGTT CTGCAAG |
| IRR | 14861-14907 | - | 47 | GGGGGCGTGC CGAGATAAGC CAAAAATTAG GACATTCGTT CTGTAAG |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res_site | Tn5041 | 13653-13686 | + | 34 | TTGACCCACG ATAAGCGCGG TTATCGTGAG TTAA |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| tnpA | Tn5041 | 151-3168 | + | Transposase | |
| tnpC | Tn5041 | 3168-3530 | + | Accessory Gene | Inhibitor/Regulator |
| urf2-ARD70099.1 | Tn5041 | 3824-4201 | - | Passenger Gene | Hypothetical |
| merE | Tn5041 | 4198-4401 | - | Passenger Gene | Heavy Metal Resistance |
| merD | Tn5041 | 4427-4789 | - | Passenger Gene | Heavy Metal Resistance |
| orfY-WP_017849615.1 | Tn5041 | 4786-5205 | - | Passenger Gene | Hypothetical |
| merA | Tn5041 | 5226-6905 | - | Passenger Gene | Heavy Metal Resistance |
| merC | Tn5041 | 6938-7372 | - | Passenger Gene | Heavy Metal Resistance |
| merP | Tn5041 | 7385-7663 | - | Passenger Gene | Heavy Metal Resistance |
| merT | Tn5041 | 7677-8027 | - | Passenger Gene | Heavy Metal Resistance |
| merR | Tn5041 | 8099-8539 | + | Passenger Gene | Heavy Metal Resistance |
| merR 3'-end | Tn5041 | 9492-9754 | + | Passenger Gene | Heavy Metal Resistance |
| oprD family | Tn5041 | 11510-11905 | + | Passenger Gene | Other |
| tnpT | Tn5041 | 12515-13564 | - | Accessory Gene | Resolvase |
| tnpS | Tn5041 | 13706-14761 | + | Accessory Gene | Resolvase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn5041 | 1005 | 151-3168 | + |
|---|
| Class: | Transposase | ||||||||||
| Function: | GO molecular function: DNA binding, transposase activity | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MASVERTAYP LLPSQLPAKE LHRCYSLSDS EIEWVNNTAK SPALSIGLAI QLKVFQQLHY FVPFEELPQE LISHVRQCLR YGARIAPRYS NPRTLYRHQA AVRQYLQVTP FYSSDGLAIT EEIARDCAVV LEQRVDLINA MLDELIQRGY ELPAYSTLNN IAETALASAQ EVTFNLIVLR APIEVIYKLK ELLDTDFGRR QSDFNALKQA PKKPSRKHLE VLIDHLAWLE SFGDLDAILE GVVDAKIRHF ATQAAASDVA ELKDCSLPKR YTLMLALIYR MRVRTRDHLA EMFIRRISTI HKRAKEEKEQ IQARQRQKLE QLAATLDGVV QILVQEPDDQ EAGSLIREFL SPDGNLDRLR ETCAEVQATG GNNYLPLIWK HFKSHRSLLF RLSHLLQLEP TTQDRSLIQA LQLIQDSENL HREWIDEHVD LSFASERWVK VVRRPASEGP PTNRRYLEVC VFSYLASELR SGDMCVLGSE SFADYRKQLL PWEECFHRLP AYCEKVGLPG TAKEFVASLK IQLEETAQHL DEKFPSCRGD VSINEAGEPV LRRVIARDIP PSAISLQTAL MQRMPARHVL DIMANIEHWI QFTRHFGPMS GNEPKLKEPA ERYLMTIFAM GCNLGPNQAA RHLAGNVTPH MLSYTNRRHL SLEKLDKANR ELVELYLQLD LPKLWGDGKA VAADGTQFDF YDDNLLAGYH FRYRKMGAVA YRHVANNYIA VFQHFIPPGI WEAIYVIEGL LKVDLSVEPD TVYSDTQGQS ATVFAFTHLL GINLMPRIRN WRDLVMCRPD RGVSYKHINR LFTDTADWHL IETHWQDLMQ VALSIQAGKI SSPMLLRKLG SYSRRNKLYH AAQALGSVIR TIFLLNWIGS RELRQEVTAN TNKIESYNGF SKWLSFGGDV IAENDPDEQQ KRLRYNDMVA SSVILQNTVD MMRILQKLAR DGWQFTDDDV SFLSPYLTSN VKRFGEFNLK LKRPPEPWIK DSIFQQAAGS MRAKQMSDSN VEVTN |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpC | TnpC | Tn5041 | 120 | 3168-3530 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Inhibitor/Regulator | ||||||||||
| Sequence Family: | Tn3_family | ||||||||||
| Comment: | putative tnpA repressor | ||||||||||
| Protein Sequence: | MHIPPASFRV TPYGDVDTKV LDSLRASYDT AQLLNLVDRL DACLAEIGGV GSIREDFLRL HGMAMTVLEG FPLTVATSDV DSIWAQAMAL QEDISALCSC LQAGSKHVAP LAALAPDYQD |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | urf2-ARD70099.1 | Urf2-ARD70099.1 | Tn5041 | 125 | 3824-4201 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Hypothetical | ||||||||||
| Function: | The EAL domain (InterPro:IPR035919) is found in diverse bacterial signalling proteins. It has been shown tostimulate degradation of a second messenger, cyclic di-GMP,and is a good candidate for a diguanylate phosphodiesterasefunction. | ||||||||||
| Protein Sequence: | MTRSEHGAVS AAELRQAWER WELLLHCQPQ VDLRAGSIAG VEELLGFLGR WRNPIMNAND TPTISCCECC KEISLDAALT PEGTEYVVHF CGLECYQRFI SRADKRAASE PDHPCTPEVS TRHDD |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merE | MerE | Tn5041 | 67 | 4198-4401 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercury ion transmembrane transporter activity (GO:0015097) | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MPRWHAYTWG VLAALTCPCH LPVLALLLSG TAAGAFVSEH WSLATLVLTV LFVLFLRAAL RAFRKRA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merD | MerD | Tn5041 | 120 | 4427-4789 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | DNA binding (GO:0003677)||negative regulation oftranscription, DNA templated (GO:0045892)||response to mercury ion (GO:0046689) | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MNAYSISRLA EDAGVSVHIV RDYMLRGLLH PARRTESGYG IFDERSLARL CFVRAAFESG IGLDELARLC RALDADDSTD VIGCIDRLLG QIATRLAALD AVKTELAGLT HRAIEGHTHA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | orfY-WP_017849615.1 | OrfY-WP_017849615.1 | Tn5041 | 139 | 4786-5205 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Hypothetical | ||||||||||
| Protein Sequence: | MSNTADDLGR CDSLRKTWQL VSFWGLPAVV ATIAIVLSNR RPALFLAAGA ALAVMGGACL VNATRCRRLH CYITGPHFLL LAVGALLAYV FDQNGEHLSR LWLLLALGAA PLLIWLPERL AGRKYLSAPV CGEGRECRR |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merA | MerA | Tn5041 | 559 | 5226-6905 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | electron transfer activity (GO:0009055)||flavin adenine dinucleotide binding (GO:0050660)||mercury (II) reductase activity (GO:0016152)||mercury ion binding (GO:0045340)||NADP binding (GO:0050661)||oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor (GO:0016668)||cell redox homeostases (GO:0045454)||detoxification of mercury ion (GO:0050787)||metal ion transport (GO:0030001) | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MTELKIIGMT CASCVVHVKE ALEKIPGVHR ADVSYASGKA ELKVDEGASR EQMQAAVEAL GYRAAFEDAM VQARPGLLDK ARGWLSSTDA GKEGDKLHVA VIGSGGAAMA AALKVVEGGA RVTLIERGII GGTCVNVGCV PSKIMIRAAH VAHLRRESPF DAGLSAMTPT VLRERLLAQQ QDRVAELRHA KYESILESTP AISVLRGTAR FQDGHTLSVK LAEGGEHIVA FDRCLVATGA SAAVPPIPGL KDTPYWTSDQ ALASDTIPKR LAVIGASVVA VELAQAFARL GSEVTILARS AMFFHEDPAI GAAVTEAFRM EGIEVLEQTQ ASQVSHANGE FVLATNHGEL RADQLLVATG RTPNTQGLNL EAADVQLDER GGIQIDERMR TSAADIYAAG DCTDQPQFVY VAAAAGTRAA INMTGGEAKL NLDVMPAVVF TDPQVATVGY SEAEAQHAGI ETDSRTLTLD NVPRALANFD TRGFIKLVAE AGSGRLLGVQ AVAPEAGELI QTAVLAIRNR MTVQELADQL FPYLTMVEGL KLAAQTFNKD VKQLSCCAG |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merC | MerC | Tn5041 | 144 | 6938-7372 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercury ion transmembrane transporter activity (GO:0015097)||metal ion binding (GO:0046872) | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MANPLNLFTR IGDKAGSVGV LISAIGCAMC FPALASLGAA FGLGFLSQWE GLFITTLLPL FAGLALLVNA LGWLNHRQWR RTALGLIGPT LVLAAVFFMR AYGWRSGWLL YIGLALMFGV SIWDLVSPAH RRCGPDACPT PPNQ |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merP | MerP | Tn5041 | 92 | 7385-7663 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercury ion binding (GO:0045340)||mercury ion transmembrane transporter activity (GO:0015097) | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MRKLLASLAL ASLVATPVWA ATQTVTLAVP GMTCAACPIT VKTALTKVDG VTKAEVSFEN REAIVTFDDT KTNALALTKA TEDAGYPSSV KQ |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merT | MerT | Tn5041 | 116 | 7677-8027 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercury ion binding (GO:0045340)||mercury ion transmembrane transporter activity (GO:0015097) | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MSEPSNGRAP LVAGGLAAIL ASACCLGPLI LIALGFSGAW IGNLTVLEPY RPLFIGAALV ALFFAYRRIF RPAQACAPGE VCAIPQVRTS YKLLYWLVAA LVLVALGFPY ILPLFY |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merR | MerR | Tn5041 | 146 | 8099-8539 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | DNA binding (GO:0003677)||mercury ion binding (GO:0045340)||regulation of transcription, DNA templated (GO:0006355)||response to mercury ion (GO:0046689) | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MKNNLESLTI GAFAKAAGVN VETIRFYQRK ALLPEPDKPY GSIRRYGEAD VARVKFVKSA QRLGFSLDEV AGLLRLDDGA HCDEARVLAE QKLGDVRGKL ADLRRIESVL EQLVHDCCAS HGTVSCPLIV SLHGDNSGVN RGFPVA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merR 3'-end | N/A | Tn5041 | 87 | 9492-9754 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Target: | Mercury | ||||||||||
| Comment: | incomplete | ||||||||||
| Protein Sequence: | DVARVKFVKS AQRLGFSLEE VAGLLRLDDG AHCDEARVLA EQKLGDVRGK LADLRRIESV LVQLVHDCCA SHGTVSCPLI VSLHG*N |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | oprD family | OprD family | Tn5041 | 131 | 11510-11905 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Sequence Family: | outer membrane porin, OprD family (Pfam:PF03573)||Porin domain superfamily (Interpro:IPR023614) | ||||||||||
| Comment: | displays sequence similarity to porins D2 and E1 described in PMID:8394980 | ||||||||||
| Protein Sequence: | MGPRSHRPIR VGLGQAEDNY SKLGGAVKTR FLDTEIKVGD VFPVTPVVQY GDSRLLPESF RGVTAYNTSV EGLVLQGGRL HAMSQPSSSS MRDDFATFYA GEVGSPWIVG GDYTPNDNLG FSLYTSCLKD A |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpT | TnpT | Tn5041 | 349 | 12515-13564 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Function: | DNA binding (GO:0003677)||DNA integration (GO:0015074)||DNA recombination (GO:0006310) | ||||||||||
| Comment: | enhances recombination (resolution)||integrase-like protein | ||||||||||
| Protein Sequence: | MARSGVLYLH VAQAATQLVA AGHNPTIDSI RVALGGTGSK STIAPLLKRW KAAHPGTLAQ AELGLPAELV LALKGLYEKV QAEAAVQLQQ AVAAHQAEAD ALQEQLQQAF VERDAQLSVQ EQQAQALAVA TTRSQGLADT LQRQEIALAS LGSEKTGLEQ RLADRAAEAA ALTRQLQQAR EQFEHYQASV AQQRSDERQA TEQRQQRLEH ELAALRQRLL AQQTRLGELQ AQEQRLVQDH DRLESTLLTT QATLAQSQIA HEHVLLQFTD LQQSHQALEQ RHGQGEQRLA ETRTYLAVNE RERSLLAERL SQTESQLSQL ATEKQLLVQD NAVLSSQLAE SRAMKPKTC |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpS | TnpS | Tn5041 | 351 | 13706-14761 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Function: | DNA binding (GO:0003677)||recombinase activity (GO:0000150)||DNA integration (GO:0015074) | ||||||||||
| Sequence Family: | Tyrosine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Tyrosine | ||||||||||
| Protein Sequence: | MSIICGTHGL NRRFVMTAGN NDENLPTRRH EEPTVLARTP GTLTTPEQLA EQHQRFLAAA TTDNTRRTYR SAIRHFLAWG GVLPCDEAAL IRYLLSFAEV LNPRTLALRL TALSQWHRYQ GFPDPTASAT AGKTLRGIER VNGRPRQKAK ALVLEDLERI VVHLNTLDGL ATLRDSALLQ VGYFGAFRRS ELVTLEMQYL EWEQEGLRIT LPRSKTDQEG EGLDKAIPYG DSICCPATAL RRWLDAAQIV QGPLFRRISR WGVLGEVALH EGSVNTILTA RAEAAGLLYV PELSSHSLRR GLATSAHRAG ADFLEIKRQG GWRHDGTVHG YIEEAGAFEE NAAGSLLRRK P |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| kappa_gamma-X98999.3 | kappa_gamma | transposon | 9230-9491 | + | 262 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRR | kappa_gamma | 9230-9267 | + | 38 | GGGGTTGGGG GAGCAACGGA ACAGAAAGTG CACTTAAG |
| IRL | kappa_gamma | 9454-9491 | - | 38 | GGGGGTTGGG GAGCAATGGA ACTGAAAACC AACGTAAG |
1. Kholodii GY, Yurieva OV, Gorlenko ZM, Mindlin SZ, Bass IA, Lomovskay OL, Kopteva AV, Nikiforov VG.
Tn5041: a chimeric mercury resistance transposon closely related to the toluene degradative transposon Tn4651.. Microbiology (Reading). 1997 Aug;143 ( Pt 8):2549-2556. doi: 10.1099/00221287-143-8-2549. PubMed ID:
9274008
.
2. Yamano Y, Nishikawa T, Komatsu Y. Cloning and nucleotide sequence of anaerobically induced porin protein E1 (OprE) of Pseudomonas aeruginosa PAO1.. Mol Microbiol. 1993 May;8(5):993-1004. doi: 10.1111/j.1365-2958.1993.tb01643.x. PubMed ID: 8394980 .
3. Kholodii GIa, Mindlin SZ, Gorlenko ZhM, Bass IA, Kaliaeva ES, Nikiforov VG. [Molecular genetic analysis of the Tn5041 transposition system].. Genetika. 2000 Apr;36(4):459-69. PubMed ID: 10822806 .
4. Kholodii G, Gorlenko Z, Mindlin S, Hobman J, Nikiforov V. Tn5041-like transposons: molecular diversity, evolutionary relationships and distribution of distinct variants in environmental bacteria.. Microbiology (Reading). 2002 Nov;148(Pt 11):3569-3582. doi: 10.1099/00221287-148-11-3569. PubMed ID: 12427948 .
2. Yamano Y, Nishikawa T, Komatsu Y. Cloning and nucleotide sequence of anaerobically induced porin protein E1 (OprE) of Pseudomonas aeruginosa PAO1.. Mol Microbiol. 1993 May;8(5):993-1004. doi: 10.1111/j.1365-2958.1993.tb01643.x. PubMed ID: 8394980 .
3. Kholodii GIa, Mindlin SZ, Gorlenko ZhM, Bass IA, Kaliaeva ES, Nikiforov VG. [Molecular genetic analysis of the Tn5041 transposition system].. Genetika. 2000 Apr;36(4):459-69. PubMed ID: 10822806 .
4. Kholodii G, Gorlenko Z, Mindlin S, Hobman J, Nikiforov V. Tn5041-like transposons: molecular diversity, evolutionary relationships and distribution of distinct variants in environmental bacteria.. Microbiology (Reading). 2002 Nov;148(Pt 11):3569-3582. doi: 10.1099/00221287-148-11-3569. PubMed ID: 12427948 .