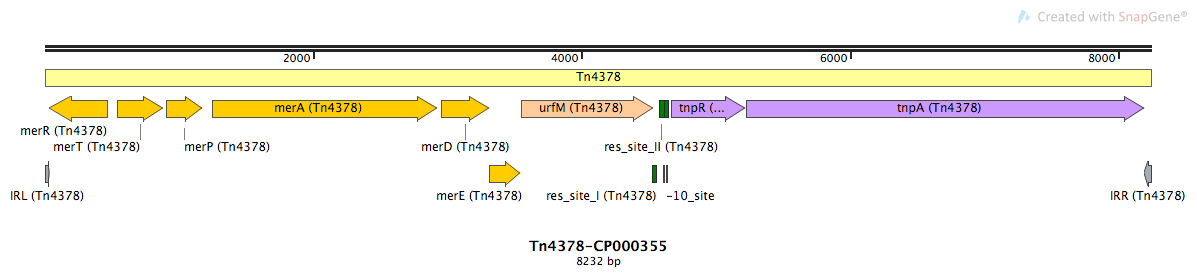
Mobile element type:
Transposon
Name:
Tn4378
Synonyms:
Accession:
Tn4378-CP000355
Family:
Tn3
Group:
Tn21
First isolate:
Yes
Partial:
ND
Evidence of transposition:
ND
Host Organism:
Cupriavidus metallidurans CH34
Date of Isolation:
Country:
Belgium
Molecular Source:
plasmid pMOL28
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-38 | + | 38 | GGGGTCGTCT CAGAAAACGG AAAATAAAGC ACGCTAAG |
| IRR | 8195-8232 | - | 38 | GGGGTCGTCT CAGAAAACGG AAAATAAAGC ACGCTAAG |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res_site_I | Tn4378 | 4527-4557 | + | 31 | CGTCAGATTG AGGCATACCC TAACCGGATG T |
| res_site_II | Tn4378 | 4578-4612 | + | 35 | CGTCAGAATA GAGTCGGTTG TGTTATTTAT TGACA |
| res_site_III | Tn4378 | 4615-4646 | + | 32 | AGCTGAAAAA GGTCATAGAT TTCTTCCTGA CA |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| merR | Tn4378 | 34-468 | - | Passenger Gene | Heavy Metal Resistance |
| merT | Tn4378 | 540-890 | + | Passenger Gene | Heavy Metal Resistance |
| merP | Tn4378 | 903-1178 | + | Passenger Gene | Heavy Metal Resistance |
| merA | Tn4378 | 1250-2935 | + | Passenger Gene | Heavy Metal Resistance |
| merD | Tn4378 | 2953-3318 | + | Passenger Gene | Heavy Metal Resistance |
| merE | Tn4378 | 3315-3551 | + | Passenger Gene | Heavy Metal Resistance |
| urfM | Tn4378 | 3548-4537 | + | Passenger Gene | Other |
| tnpR | Tn4378 | 4667-5227 | + | Accessory Gene | Resolvase |
| tnpA | Tn4378 | 5230-8199 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merR | MerR | Tn4378 | 144 | 34-468 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | activator-repressor of mer operon | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MENNLENLTI GVFAKAAGVN VETIRFYQRK GLLLEPDKPY GSIRRYGEAD VTRVRFVKSA QRLGFSLDEI AELLRLEDGT HCEEASSLAE HKLKDVREKM ADLARMEAVL SELVCACHAR RGNVSCPLIA SLQGGASLAG SAMP |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merT | MerT | Tn4378 | 116 | 540-890 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | cytosolic mercuric ion transport protein | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MSEPKTGRGA LFTGGLAAIL ASACCLGPLV LIALGFSGAW IGNLAVLDPY RPIFIGVALV ALFFAWRRIY RQAAACKPGE VCAIPQVRAT YKLIFWIVAA LVLVALGFPY VMPFFY |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merP | MerP | Tn4378 | 91 | 903-1178 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercury transport | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MKKLFASLAL AAVVAPVWAA TQTVTLSVPG MTCSACPITV KKAISKVEGV SKVDVTFETR QAVVTFDDAK TSVQKLTKAT ADAGYPSSVK Q |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merA | MerA | Tn4378 | 561 | 1250-2935 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercuric ion reductase | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MTHLKITGMT CDSCAAHVKE ALEKVPGVQS ALVSYPKGTA QLAIVPGTSP DALTAAVAGL GYKATLADAP LADNRVGLLD KVRGWMAAAE KHSGNEPPVQ VAVIGSGGAA MAAALKAVEQ GAQVTLIERG TIGGTCVNVG CVPSKIMIRA AHIAHLRRES PFDGGIAATV PTIDRSKLLA QQQARVDELR HAKYEGILGG NPAITVVHGE ARFKDDQSLT VRLNEGGERV VMFDRCLVAT GASPAVPPIP GLKESPYWTS TEALASDTIP ERLAVIGSSV VALELAQAFA RLGSKVTVLA RNTLFFREDP AIGEAVTAAF RAEGIEVLEH TQASQVAHMD GEFVLTTTHG ELRADKLLVA TGRTPNTRSL ALDAAGVTVN AQGAIAIDQG MRTSNPNIYA AGDCTDQPQF VYVAAAAGTR AAINMTGGDA ALDLTAMPAV VFTDPQVATV GYSEAEAHHD GIETDSRTLT LDNVPRALAN FDTRGFIKLV IEEGSHRLIG VQAVAPEAGE LIQTAALAIR NRMTVQELAD QLFPYLTMVE GLKLAAQTFN KDVKQLSCCA G |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merD | MerD | Tn4378 | 121 | 2953-3318 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | secondary regulatory protein | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MNAYTVSRLA LDAGVSVHIV RDYLLRGLLR PVACTPGGYG LFDDAALQRL CFVRAAFEAG IGLDALARLC RALDAADGDE AAAQLALLRQ FVERRREALA DLEVQLATLP TEPAQHAESL P |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merE | MerE | Tn4378 | 78 | 3315-3551 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercury transport | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MNNPERLPSE THKPITGYLW GGLAVLTCPC HLPILAVVLA GTTAGAFLGE HWVIAALGLT GLFLLSLSRA LRAFRERE |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | urfM | UrfM | Tn4378 | 329 | 3548-4537 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Function: | possible diguanylate phosphodiesterase | ||||||||||
| Sequence Family: | EAL (Pfam:PF00563)||DUF3330 (Pfam:PF11809) | ||||||||||
| Comment: | similar to urfM from E.coli | ||||||||||
| Protein Sequence: | MSAFRPDGWT TPELAQAVER GQLELHYQPV VDLRSGGIVG AEALLRWRHP TLGLLPPGQF LPVVESSGLM PEIGAWVLGE ACRQMRDWRM LAWRPFRLAV NVSASQVGPD FDGWVKGVLA DAELPAEYLE IELTESVAFG DPAIFPALDA LRQIGVRFAA DDFGTGYSCL QHLKCCPIST LKIDQSFVAG LANDRRDQTI VHTVIQLAHG LGMDVVAEGV ETSASLDLLR QADCDTGQGF LFAKPMPAAA FAVFVSQWRG ATMNASDSTT TSCCVCCKEI PLDAAFTPEG AEYVEHFCGL ECYQRFEARA KTGNETDADP NACDSLPSD |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn4378 | 186 | 4667-5227 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Function: | resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MQGQRIGYVR VSSFDQNPER QLEHVEVGKV FTDKASGKDT QRPELDSLLA FVREGDTVVV HSMDRLARNL DDLRRLVQKL TKRGVRIEFV KESLTFTGED SPMANLMLSV MGAFAEFERA LIRERQREGI ALAKQRGAYR GRKKALSPEQ VADLRQRAAA GEQKAKLARE FGVSRETLYQ YLRADQ |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn4378 | 989 | 5230-8199 | + |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MPRRSILSAA ERESLLALPD TKDDLIRYYT FSDTDLSIIR QRRGPANRLG FAVQLCYLRF PGILLGVDEP PFPPLLKLVA DQLKVSVESW GEYGQREQTR REHLVELQTV FGFQPFTMSH YRQAVHTLTE LAMQTDKGIV LASALIEHLR RQSVILPALN AVERASAEAI TRANRRIYYA LAEPLSDAHR RRLDDLLKRR DNGKTTWLAW LRQSPVKPNS RHMLEHIERL KAWQALDLPT GIERLIHQNR LLKIAREGGQ MTPADLAKFE AQRRYATLVA LAIEGMATVT DEIIDLHDRI LGKLFNAAKN KHQQQFQASG KAINAKVRLF GRIGQALIEA KQSGRDPFAA IEAVMSWDAF AESVTEAQKL AQPEDFDFLH RIGESYATLR RYAPEFLAVL KLRAAPAAKD VLEAIEVLRN MNSDNARKVP ADAPTDFIKP RWQKLVMTDT GIDRRYYELC ALSEMKNALR SGDIWVQGSR QFKDFEDYLV PPAKFASLKQ ASELPLAVAT DCDQYLHDRL TLLETQLATV NRMALANELP DAIITESGLK ITPLDAAVPD TAQALIDQTA MILPHVKITE LLLEVDEWTG FTRHFAHLKS GDLAKDKNLL LTTILADAIN LGLTKMAESC PGTTYAKLAW LQAWHTRDET YSSALAELVN AQFRHPFAEH WGDGTTSSSD GQNFRTGSKA ESTGHINPKY GSSPGRTFYT HISDQYAPFH TKVVNVGVRD STYVLDGLLY HESDLRIEEH YTDTAGFTDH VFGLMHLLGF RFAPRIRDLG DTKLFIPKGD TVYDALKPMI SSDRLNIKAI RAHWDEILRL ATSIKQGTVT ASLMLRKLGS YPRQNGLAVA LRELGRIERT LFILDWLQSV ELRRRVHAGL NKGEARNALA RAVFFNRLGE IRDRSFEQQR YRASGLNLVT AAVVLWNTVY LERAAHALRG NGHAVDDALL QYLSPLGWEH INLTGDYLWR SSAKIGAGKF RPLRPLQPA |
||||||||||
1. Taghavi S, Mergeay M, van der Lelie D.
Genetic and physical maps of the Alcaligenes eutrophus CH34 megaplasmid pMOL28 and its derivative pMOL50 obtained after temperature-induced mutagenesis and mortality.. Plasmid. 1997;37(1):22-34. doi: 10.1006/plas.1996.1274. PubMed ID:
9073579
.
2. Van Houdt R, Monchy S, Leys N, Mergeay M. New mobile genetic elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria.. Antonie Van Leeuwenhoek. 2009 Aug;96(2):205-26. doi: 10.1007/s10482-009-9345-4. Epub 2009 Apr 24. PubMed ID: 19390985 .
2. Van Houdt R, Monchy S, Leys N, Mergeay M. New mobile genetic elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria.. Antonie Van Leeuwenhoek. 2009 Aug;96(2):205-26. doi: 10.1007/s10482-009-9345-4. Epub 2009 Apr 24. PubMed ID: 19390985 .