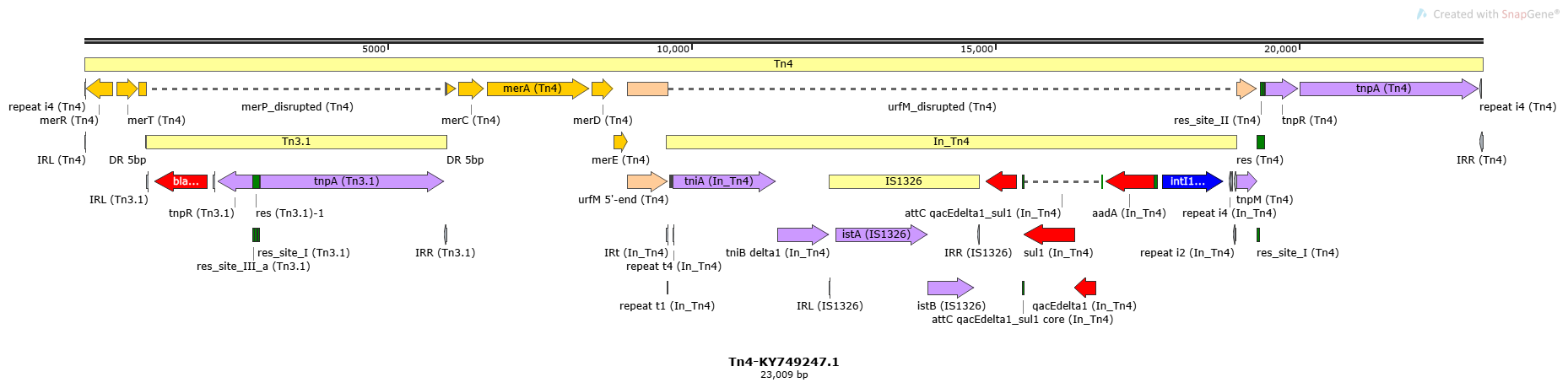
Mobile element type:
Transposon
Name:
Tn4
Synonyms:
Accession:
Tn4-KY749247.1
Family:
Tn3
Group:
Tn21
First isolate:
ND
Partial:
ND
Evidence of transposition:
ND
Host Organism:
Salmonella enterica subsp. enterica serovar Paratyphi B
Date of Isolation:
Country:
Molecular Source:
plasmid R1
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-38 | + | 38 | GGGGGCACCT CAGAAAACGG AAAATAAAGC ACGCTAAG |
| repeat i4 | 10-28 | + | 19 | TCAGAAAACG GAAAATAAA |
| IRR | 22972-23009 | - | 38 | GGGGTCGTCT CAGAAAACGG AAAATAAAGC ACGCTAAG |
| repeat i4 | 22982-23000 | - | 19 | TCAGAAAACG GAAAATAAA |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res | Tn3.1 | 2769-2889 | + | 121 | AAATGTACCT TAAATCGAAT ATCAGACACG ATGTGTCTAT TATGCCAAAA TGACGATTTA ATGGACACTC AAACGAAGCC GTTTTACTAT GTCTGATAAT TTATAATATT TCGAACGGTT G |
| res_site_III_a | Tn3.1 | 2772-2794 | + | 23 | TGTACCTTAA ATCGAATATC AGA |
| res_site_II_a | Tn3.1 | 2800-2835 | + | 36 | TGTGTCTATT ATGCCAAAAT GACGATTTAA TGGACA |
| res_site_I | Tn3.1 | 2858-2886 | + | 29 | TGTCTGATAA TTTATAATAT TTCGAACGG |
| attC qacEdelta1_sul1 | In_Tn4 | 15443-15476 , 16754-16760 | + | 41 | CCGCTAGCGG GCGGCCGGAA GGTGAATGCT AGGCATCTAA C |
| attC qacEdelta1_sul1 core | In_Tn4 | 15443-15476 | + | 34 | CCGCTAGCGG GCGGCCGGAA GGTGAATGCT AGGC |
| attI | In_Tn4 | 17616-17671 | + | 56 | CTTTGTTTTA GGGCGACTGC CCTGCTGCGT AACATCGTTG CTGCTCCATA ACATCA |
| res | Tn4 | 19306-19436 | + | 131 | GCCGCCGTCA GGTTGAGGCA TACCCTAACC TGATGTCAGA TGCCATGTGT AAATTGCGTC AGGATAGGAT TGAATTTTGA ATTTATTGAC ATATCTCGTT GAAGGTCATA GAGTCTTCCC TGACATTTTG C |
| res_site_I | Tn4 | 19306-19344 | + | 39 | GCCGCCGTCA GGTTGAGGCA TACCCTAACC TGATGTCAG |
| res_site_II | Tn4 | 19358-19401 | + | 44 | ATTGCGTCAG GATAGGATTG AATTTTGAAT TTATTGACAT ATCT |
| res_minus_35 | Tn4 | 19391-19396 | + | 6 | TTGACA |
| res_site_III | Tn4 | 19405-19436 | + | 32 | TGAAGGTCAT AGAGTCTTCC CTGACATTTT GC |
| res_minus_35 | Tn4 | 19563-19568 | - | 6 | TTGACA |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| merR | Tn4 | 34-468 | - | Passenger Gene | Heavy Metal Resistance |
| merT | Tn4 | 540-890 | + | Passenger Gene | Heavy Metal Resistance |
| merP | Tn4 | 904-1019 , 5966-6132 | + | Passenger Gene | Heavy Metal Resistance |
| merP 5'-end | Tn4 | 904-1019 | + | Passenger Gene | Heavy Metal Resistance |
| bla TEM-1 (ARO:3000873) | Tn3.1 | 1166-2026 | - | Passenger Gene | Antibiotic Resistance |
| tnpR | Tn3.1 | 2209-2766 | - | Accessory Gene | Resolvase |
| tnpA | Tn3.1 | 2895-5933 | + | Transposase | |
| merP 3'-end | Tn4 | 5966-6132 | + | Passenger Gene | Heavy Metal Resistance |
| merC | Tn4 | 6168-6590 | + | Passenger Gene | Heavy Metal Resistance |
| merA | Tn4 | 6642-8336 | + | Passenger Gene | Heavy Metal Resistance |
| merD | Tn4 | 8354-8716 | + | Passenger Gene | Heavy Metal Resistance |
| merE | Tn4 | 8713-8949 | + | Passenger Gene | Heavy Metal Resistance |
| urfM 5'-end | Tn4 | 8946-9616 | + | Passenger Gene | Other |
| urfM 5'-end | Tn4 | 8946-9616 | + | Passenger Gene | Other |
| tniA | In_Tn4 | 9692-11407 | + | Transposase | |
| tniB delta1 | In_Tn4 | 11410-12270 | + | Accessory Gene | |
| istA | IS1326 | 12372-13895 | + | Transposase | None |
| istB | IS1326 | 13882-14667 | + | Accessory Gene | ATPase Transposition Helper |
| GNAT_fam | In_Tn4 | 14843-15343 | - | Passenger Gene | Antibiotic Resistance |
| sul1 (ARO:3000410) | In_Tn4 | 15471-16310 | - | Passenger Gene | Antibiotic Resistance |
| qacEdelta1 (ARO:3005010) | In_Tn4 | 16304-16651 | - | Passenger Gene | Antibiotic Resistance |
| aadA (ARO:3002601) | In_Tn4 | 16815-17606 | - | Passenger Gene | Antibiotic Resistance |
| intI1 | In_Tn4 | 17755-18768 | + | Integron Integrase | Class 1 |
| tnpM | Tn4 | 18971-19321 | + | Accessory Gene | Inhibitor |
| tnpR | Tn4 | 19447-20007 | + | Accessory Gene | Resolvase |
| tnpA | Tn4 | 20010-22976 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merR | MerR | Tn4 | 144 | 34-468 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | activator-repressor of mer operon | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MENNLENLTI GVFAKAAGVN VETIRFYQRK GLLREPDKPY GSIRRYGEAD VVRVKFVKSA QRLGFSLDEI AELLRLDDGT HCEEASSLAE HKLKDVREKM ADLARMETVL SELVCACHAR KGNVSCPLIA SLQGEAGLAR SAMP |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merT | MerT | Tn4 | 116 | 540-890 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | cytosolic mercuric ion transport protein | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MSEPQNGRGA LFAGGLAAIL ASTCCLGPLV LVALGFSGAW IGNLTVLEPY RPLFIGAALV ALFFAWKRIY RPVQACKPGE VCAIPQVRAT YKLIFWIVAV LVLVALGFPY VVPFFY |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merP | None | Tn4 | 94 | 904-1019 , 5966-6132 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MKKLFASLAL AAAVAPVWAA TQTVTLAVPG MTCAACPISD HSQESALQGR RREQGRCGLR EARGRRHF*R HQGQRTEADQ GHRRRRLSVQ RQAV |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merP 5'-end | N/A | Tn4 | 38 | 904-1019 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Target: | Mercury | ||||||||||
| Comment: | merP interrupted by insertion of Tn3.1 | ||||||||||
| Protein Sequence: | MKKLFASLAL AAAVAPVWAA TQTVTLAVPG MTCAACPI |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | bla TEM-1 (ARO:3000873) | Bla TEM-1 | Tn3.1 | 286 | 1166-2026 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | TEM beta-lactamase (ARO:3000014) | ||||||||||
| Target: | penem (ARO:3003706)||cephalosporin (ARO:0000032)||monobactam (ARO:0000004)||penam (ARO:3000008) | ||||||||||
| Comment: | perfect match to reference sequence for ARO:3000873||Synonyms: TEM-98, RTEM-1 | ||||||||||
| Protein Sequence: | MSIQHFRVAL IPFFAAFCLP VFAHPETLVK VKDAEDQLGA RVGYIELDLN SGKILESFRP EERFPMMSTF KVLLCGAVLS RVDAGQEQLG RRIHYSQNDL VEYSPVTEKH LTDGMTVREL CSAAITMSDN TAANLLLTTI GGPKELTAFL HNMGDHVTRL DRWEPELNEA IPNDERDTTM PAAMATTLRK LLTGELLTLA SRQQLIDWME ADKVAGPLLR SALPAGWFIA DKSGAGERGS RGIIAALGPD GKPSRIVVIY TTGSQATMDE RNRQIAEIGA SLIKHW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn3.1 | 185 | 2209-2766 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Function: | resolvase||serine site-specific recombinase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Comment: | first defined as a repressor | ||||||||||
| Protein Sequence: | MRIFGYARVS TSQQSLDIQI RALKDAGVKA NRIFTDKASG SSTDREGLDL LRMKVEEGDV ILVKKLDRLG RDTADMIQLI KEFDAQGVAV RFIDDGISTD GDMGQMVVTI LSAVAQAERR RILERTNEGR QEAKLKGIKF GRRRTVDRNV VLTLHQKGTG ATEIAHQLSI ARSTVYKILE DERAS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn3.1 | 1012 | 2895-5933 | + |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | In frame three amino acid deletion relative to tnpA (Tn3) | ||||||||||
| Protein Sequence: | VLKKPSGREA DMPVDFLTTE QVESYGRFTG EPDELQLARY FHLDEADKEF IGKSRGDHNR LGIALQIGCV RFLGTFLTDM NHIPSGVRHF TARQLGIRDI TVLAEYGQRE NTRREHAALI RQHYQYREFA WPWTFRLTRL LYTRSWISNE RPGLLFDLAT GWLMQHRIIL PGATTLTRLI SEVREKATLR LWNKLALIPS AEQRSQLEML LGPTDCSRLS LLESLKKGPV TISGPAFNEA IERWKTLNDF GLHAENLSTL PAVRLKNLAR YAGMTSVFNI ARMSPQKRMA VLVAFVLAWE TLALDDALDV LDAMLAVIIR DARKIGQKKR LRSLKDLDKS ALALASACSY LLKEETPDES IRAEVFSYIP RQKLAEIITL VREIARPSDD NFHDEMVEQY GRVRRFLPHL LNTVKFSSAP AGVTTLNACD YLSREFSSRR QFFDDAPTEI ISQSWKRLVI NKEKHITRRG YTLCFLSKLQ DSLRRRDVYV TGSNRWGDPR ARLLQGADWQ ANRIKVYRSL GHPTDPQEAI KSLGHQLDSR YRQVAARLGE NEAVELDVSG PKPRLTISPL ASLDEPDSLK RLSKMISDLL PPVDLTELLL EINAHTGFAD EFFHASEASA RVDDLPVSIS AVLMAEACNI GLEPLIRSNV PALTRHRLNW TKANYLRAET ITSANARLVD FQATLPLAQI WGGGEVASAD GMRFVTPVRT INAGPNRKYF GNNRGITWYN FVSDQYSGFH GIVIPGTLRD SIFVLEGLLE QETGLNPTEI MTDTAGASDL VFGLFWLLGY QFSPRLADAG ASVFWRMDHD ADYGVLNDIA RGQSDPRKIV LQWDEMIRTA GSLKLGKVQA SVLVRSLLKS ERPSGLTQAI IEVGRINKTL YLLNYIDDED YRRRILTQLN RGESRHAVAR AICHGQKGEI RKRYTDGQED QLGALGLVTN AVVLWNTIYM QAALDHLRAQ GETLNDEDIA RLSPLCHGHI NMLGHYSFTL AELVTKGHLR PLKEASEAEN VA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merP 3'-end | N/A | Tn4 | 55 | 5966-6132 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Target: | Mercury | ||||||||||
| Comment: | merP interrupted by insertion of Tn3.1 | ||||||||||
| Protein Sequence: | RSQSRKRSPR SKA*ARSMWA SRSARPSSLL TTPRPAYRS* PRPPQTPAIR PASSS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merC | MerC | Tn4 | 140 | 6168-6590 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | transmembrane protein mercury transport | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MGLMTRIADK TGALGSVVSA MGCAACFPAL ASFGAAIGLG FLSQYEGLFI SRLLPLFAAL AFLANALGWF SHRQWLRSLL GMIGPAIVFA ATVWLLGNWW TANLMYVGLA LMIGVSIWDF VSPAHRRCGP DGCELPAKRL |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merA | MerA | Tn4 | 564 | 6642-8336 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercuric ion reductase | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MSTLKITGMT CDSCAVHVKD ALEKVPGVQS ADVSYAKGSA KLAIEVGTSP DALTAAVAGL GYRATLADAP SVSTPGGLLD KMRDLLGRND KTGSSGALHI AVIGSGGAAM AAALKAVEQG ARVTLIERGT IGGTCVNVGC VPSKIMIRAA HIAHLRRESP FDGGIAATTP TIQRTALLAQ QQARVDELRH AKYEGILEGN PAITVLHGSA RFKDNRNLIV QLNDGGERVV AFDRCLIATG ASPAVPPIPG LKDTPYWTST EALVSETIPK RLAVIGSSVV ALELAQAFAR LGAKVTILAR STLFFREDPA IGEAVTAAFR MEGIEVREHT QASQVAYING EGDGEFVLTT AHGELRADKL LVATGRAPNT RKLALDATGV TLTPQGAIVI DPGMRTSVEH IYAAGDCTDQ PQFVYVAAAA GTRAAINMTG GDAALNLTAM PAVVFTDPQV ATVGYSEAEA HHDGIKTDSR TLTLDNVPRA LANFDTRGFI KLVVEEGSGR LIGVQAVAPE AGELIQTAAL AIRNRMTVQE LADQLFPYLT MVEGLKLAAQ TFNKDVKQLS CCAG |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merD | MerD | Tn4 | 120 | 8354-8716 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | secondary regulatory protein | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MSAYTVSQLA HNAGVSVHIV RDYLVRGLLR PVACTTGGYG VFDDAALQRL CFVRAAFEAG IGLDALARLC RALDAADGAQ AAAQLAVLRQ LVERRRAALA HLDAQLASMP AERAHEEALP |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merE | MerE | Tn4 | 78 | 8713-8949 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercury transport | ||||||||||
| Target: | Mercury | ||||||||||
| Comment: | similar to urf-1 in pKLH2 (GenBank AF213017), pKLH272 (Genbank Y08992), pMER610 (GenBank Y08993), pKLH210 (GenBank Y10102), Tn5036 (Genbank Y09025), orf1 in Tn501 (GenBank Z00027), and urf-1 in Tn5041 (GenBank X98999) | ||||||||||
| Protein Sequence: | MNAPDKLPPE TRQPVSGYLW GALAVLTCPC HLPILAAVLA GTTAGAFLGE HWGVAALALT GLFVLAVTRL LRAFRGGS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | urfM 5'-end | N/A | Tn4 | 223 | 8946-9616 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Comment: | urfMORF interrupted by insertion of In2 | ||||||||||
| Protein Sequence: | MTSSQPAGWT AAELAQAAAR GQLDLHYQPL VDLRDHRIAG AEALMRWRHP RLGLLPPGQF LPLAESFGLM PEIGAWVLGE ACRQMHKWQG PAWQPFRLAI NVSASQVGPT FDDEVKRVLA DMALPAELLE IELTESVAFG NPALFASFDA LRAIGVRFAA DDFGTGYSCL QHLKCCPITT LKIDQSFVAR LPDDARDQTI VRAVIQLAHG LGMDVIFRRR LHQ |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | urfM 5'-end | N/A | Tn4 | 223 | 8946-9616 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Comment: | urfMORF interrupted by insertion of In2 | ||||||||||
| Protein Sequence: | MTSSQPAGWT AAELAQAAAR GQLDLHYQPL VDLRDHRIAG AEALMRWRHP RLGLLPPGQF LPLAESFGLM PEIGAWVLGE ACRQMHKWQG PAWQPFRLAI NVSASQVGPT FDDEVKRVLA DMALPAELLE IELTESVAFG NPALFASFDA LRAIGVRFAA DDFGTGYSCL QHLKCCPITT LKIDQSFVAR LPDDARDQTI VRAVIQLAHG LGMDVIFRRR LHQ |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniA | TniA | In_Tn4 | 571 | 9692-11407 | + |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MLNTRVHQSE VSMATDTPRI PEQGVATLPD EAWERARRRA EIISPLAQSE TVGHEAADMA AQALGLSRRQ VYVLIRRARQ GSGLVTDLVP GQSGGGKGKG RLPEPVERVI HELLQKRFLT KQKRSLAAFH REVTQVCKAQ KLRVPARNTV ALRIASLDPR KVIRRREGQD AARDLQGVGG EPPAVTAPLE QVQIDHTVID LIVVDDRDRQ PIGRPYLTLA IDVFTRCVLG MVVTLEAPSA VSVGLCLVHV ACDKRPWLEG LNVEMDWQMS GKPLLLYLDN AAEFKSEALR RGCEQHGIRL DYRPLGQPHY GGIVERIIGT AMQMIHDELP GTTFSNPDQR GDYDSENKAA LTLRELERWL TLAVGTYHGS VHNGLLQPPA ARWAEAVARV GVPAVVTRAT SFLVDFLPIL RRTLTRTGFV IDHIHYYADA LKPWIARRER WPSFLIRRDP RDISRIWVLE PEGQHYLEIP YRTLSHPAVT LWEQRQALAK LRQQGREQVD ESALFRMIGQ MREIVTSAQK ATRKARRDAD RRQHLKTSAR PDKPVPPDTD IADPQADNLP PAKPFDQIEE W |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniB delta1 | TniB delta1 | In_Tn4 | 286 | 11410-12270 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Function: | probable ATP-binding protein. | ||||||||||
| Comment: | probably truncated by insertion of IS1326::IS1353 | ||||||||||
| Protein Sequence: | MDEYPIIDLS HLLPAAQGLA RLPADERIQR LRADRWIGYP RAVEALNRLE ALYAWPNKQR MPNLLLVGPT NNGKSMIVEK FRRTHPASSD ADQEHIPVLV VQMPSEPSVI RFYVALLAAM GAPLRPRPRL PEMEQLALAL LRKVGVRMLV IDELHNVLAG NSVNRREFLN LLRFLGNELR IPLVGVGTRD AYLAIRSDDQ LENRFEPMML PVWEANDDCC SLLASFAASL PLRRPSPIAT LDMARYLLTR SEGTIGELAH LLMAAAIVAV ESGEEAINHR TLSMAC |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | istA | IstA | IS1326 | 507 | 12372-13895 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MINVAILSAI RRWHFRDGAS IREIARRSGL SRNTVRKYLQ SKVVEPQYPA RDSVGKLSPF EPKLRQWLST EHKKTKKLRR NLRSMYRDLV ALGFTGSYDR VCAFARQWKD SEQFKAQTSG KGCFIPLRFA CGEAFQFDWS EDFARIAGKQ VKLQIAQFKL AHSRAFVLRA YYQQKHEMLF DAHWHAFQIF GGIPKRGIYD NMKTAVDSVG RGKERRVNQR FTAMVSHYLF DAQFCNPASG WEKGQIEKNV QDSRQRLWQG APDFQSLADL NVWLEHRCKA LWSELRHPEL DQTVQEAFAD EQGELMALPN AFDAFVEQTK RVTSTCLVHH EGNRYSVPAS YANRAISLRI YADKLVMAAE GQHIAEHPRL FGSGHARRGH TQYDWHHYLS VLQKKPGALR NGAPFAELPP AFKKLQSILL QRPGGDRDMV EILALVLHHD EGAVLSAVEL ALECGKPSKE HVLNLLGRLT EEPPPKPIPI PKGLRLTLEP QANVNRYDSL RRAHDAA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | istB | IstB | IS1326 | 261 | 13882-14667 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | ATPase Transposition Helper | ||||||||||
| Function: | stimulates transposition | ||||||||||
| Protein Sequence: | MMQHEGHVRI LKSLKLFGMA HAIEELGNQN SPAFNQALPM LDSLIKAEVA EREVRSVNYQ LRVAKFPVYR DLVGFDFSQS LVNEATVKQL HRCDFMEQAQ NVVLIGGPGT GKTHLATAIG TQAVMHLNRR VRFFSTVDLV NALEQEKSSG RQGQIANRLL YADLVILDEL GYLPFSQTGG ALLFHLLSKL YEKTSVILTT NLSFSEWSRV FGDEKMTTAL LDRLTHHCHI LETGNESYRF KHSSTQNKQE EKQTRKLKIE T |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | GNAT_fam | GNAT_fam | In_Tn4 | 166 | 14843-15343 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Sequence Family: | Acetyltransf_1 (Pfam:PF00583) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | putative acetyltransferase ADU64769.1 | ||||||||||
| Protein Sequence: | MDSEEPPNVR VACSGDIDEV VRLMHDAAAW MSAKGTPAWD VARIDRTFAE TFVLRSELLV ASCSDGIVGC CTLSAEDPEF WPDALKGEAA YLHKLAVRRT HAGRGVSSAL IEACRHAART QGCAKLRLDC HPNLRGLYER LGFTHVDTFN PGWDPTFIAE RLELEI |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | sul1 (ARO:3000410) | Sul1 | In_Tn4 | 279 | 15471-16310 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic target replacement (ARO:0001002) | ||||||||||
| Sequence Family: | sulfonamide resistant sul (ARO:3004238) | ||||||||||
| Transposase Chemistry: | dihydropteroate synthase | ||||||||||
| Target: | sulfonamide antibiotic (ARO:3000282)||sulfone antibiotic (ARO:3003401) | ||||||||||
| Comment: | perfect match to reference sequence for ARO:3000410 | ||||||||||
| Protein Sequence: | MVTVFGILNL TEDSFFDESR RLDPAGAVTA AIEMLRVGSD VVDVGPAASH PDARPVSPAD EIRRIAPLLD ALSDQMHRVS IDSFQPETQR YALKRGVGYL NDIQGFPDPA LYPDIAEADC RLVVMHSAQR DGIATRTGHL RPEDALDEIV RFFEARVSAL RRSGVAADRL ILDPGMGFFL SPAPETSLHV LSNLQKLKSA LGLPLLVSVS RKSFLGATVG LPVKDLGPAS LAAELHAIGN GADYVRTHAP GDLRSAITFS ETLAKFRSRD ARDRGLDHA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | qacEdelta1 (ARO:3005010) | QacEdelta1 | In_Tn4 | 115 | 16304-16651 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic efflux (ARO:0010000) | ||||||||||
| Sequence Family: | major facilitator superfamily (MFS) antibiotic efflux pump (ARO:0010002) | ||||||||||
| Target: | disinfecting agents and antiseptics(ARO:3005386) | ||||||||||
| Comment: | subunit of the qac multidrug efflux pump||perfect match to reference sequence for ARO:3005010 (bitscore:219) | ||||||||||
| Protein Sequence: | MKGWLFLVIA IVGEVIATSA LKSSEGFTKL APSAVVIIGY GIAFYFLSLV LKSIPVGVAY AVWSGLGVVI ITAIAWLLHG QKLDAWGFVG MGLIIAAFLL ARSPSWKSLR RPTPW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | aadA (ARO:3002601) | AadA | In_Tn4 | 263 | 16815-17606 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | ANT(3'')(ARO:3004275) | ||||||||||
| Transposase Chemistry: | aminoglycoside nucleotidyltransferase | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | perfect match to reference sequence for ARO:3002601||Synonyms: aadA1-pm, aadA, aadA1, aad(3'')(9) | ||||||||||
| Protein Sequence: | MREAVIAEVS TQLSEVVGVI ERHLEPTLLA VHLYGSAVDG GLKPHSDIDL LVTVTVRLDE TTRRALINDL LETSASPGES EILRAVEVTI VVHDDIIPWR YPAKRELQFG EWQRNDILAG IFEPATIDID LAILLTKARE HSVALVGPAA EELFDPVPEQ DLFEALNETL TLWNSPPDWA GDERNVVLTL SRIWYSAVTG KIAPKDVAAD WAMERLPAQY QPVILEARQA YLGQEEDRLA SRADQLEEFV HYVKGEITKV VGK |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | intI1 | IntI1 | In_Tn4 | 337 | 17755-18768 | + |
|---|
| Class: | Integron Integrase | ||||||||||
| Subclass: | Class 1 | ||||||||||
| Sequence Family: | Class 1 Integron Tyrosine Integrase | ||||||||||
| Transposase Chemistry: | Tyrosine | ||||||||||
| Protein Sequence: | MKTATAPLPP LRSVKVLDQL RERIRYLHYS LRTEQAYVHW VRAFIRFHGV RHPATLGSSE VEAFLSWLAN ERKVSVSTHR QALAALLFFY GKVLCTDLPW LQEIGRPRPS RRLPVVLTPD EVVRILGFLE GEHRLFAQLL YGTGMRISEG LQLRVKDLDF DHGTIIVREG KGSKDRALML PESLAPSLRE QLSRARAWWL KDQAEGRSGV ALPDALERKY PRAGHSWPWF WVFAQHTHST DPRSGVVRRH HMYDQTFQRA FKRAVEQAGI TKPATPHTLR HSFATALLRS GYDIRTVQDL LGHSDVSTTM IYTHVLKVGG AGVRSPLDAL PPLTSER |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpM | TnpM | Tn4 | 116 | 18971-19321 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Inhibitor | ||||||||||
| Function: | transposition regulator||reported to enhance Tn21 transposition and suppress resolution of cointegrate replicons in vivo | ||||||||||
| Comment: | 3'-end of urfM ORF, which is interrupted by insertion of In2||inhibits tranposition probably by inhibiting resolution | ||||||||||
| Protein Sequence: | MEVVAEGVET PDCLAWLRQA GCDTVQGFLF ARPMPAAAFV GFVNQWRNTT MNANEPSTSC CVCCKEIPLD AAFTPEGAEY VEHFCGLECY QRFQARASTA TETSVKPDAC DSPPSG |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn4 | 186 | 19447-20007 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Function: | resolvase||serine site-specific recombinase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Comment: | identical to tnpR (TnAs3) | ||||||||||
| Protein Sequence: | MTGQRIGYIR VSTFDQNPER QLEGVKVDRA FSDKASGKDV KRPQLEALIS FARTGDTVVV HSMDRLARNL DDLRRIVQTL TQRGVHIEFV KEHLSFTGED SPMANLMLSV MGAFAEFERA LIRERQREGI ALAKQRGAYR GRKKSLSSER IAELRQRVEA GEQKTKLARE FGISRETLYQ YLRTDQ |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn4 | 988 | 20010-22976 | + |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | identical to TnAs3 tnpA | ||||||||||
| Protein Sequence: | MPRRSILSAA ERESLLALPD SKDDLIRHYT FNDTDLSIIR QRRGPANRLG FAVQLCYLRF PGVILGVDEL PFPPLLKLVA DQLKVGVESW NEYGQREQTR REHLSELQTV FGFRPFTMSH YRQAVQMLTE LAMQTDKGIV LASALIGHLR RQSVILPALN AVERASAEAI TRANRRIYDA LAEPLADAHR RRLDDLLKRR DNGKTTWLAW LRQSPAKPNS RHMLEHIERL KAWQALDLPT GIERLVHQNR LLKIAREGGQ MTPADLAKFE PQRRYATLVA LATEGMATVT DEIIDLHDRI LGKLFNAAKN KHQQQFQASG KAINAKVRLY GRIGQALIDA KQSGRDAFAA IEAVMSWDSF AESVTEAQKL AQPDDFDFLH RIGESYATLR RYAPEFLAVL KLRAAPAAKN VLDAIEVLRG MNTDNARKLP ADAPTGFIKP RWQKLVMTDA GIDRRYYELC ALSELKNSLR SGDIWVQGSR QFKDFEDYLV PPEKFTSLKQ SSELPLAVAT DCEQYLHERL TLLEAQLATV NRMAAANDLP DAIITESGLK ITPLDAAVPD TAQALIDQTA MVLPHVKITE LLLEVDEWTG FTRHFTHLKS GDLAKDKNLL LTTILADAIN LGLTKMAESC PGTTYAKLAW LQAWHTRDET YSTALAELVN AQFRHPFAGH WGDGTTSSSD GQNFRTASKA KSTGHINPKY GSSPGRTFYT HISDQYAPFH TKVVNVGLRD STYVLDGLLY HESDLRIEEH YTDTAGFTDH VFALMHLLGF RFAPRIRDLG DTKLYIPKGD AAYDALKPMI GGTLNIKHVR AHWDEILRLA TSIKQGTVTA SLMLRKLGSY PRQNGLAVAL RELGRIERTL FILDWLQSVE LRRRVHAGLN KGEARNALAR AVFFNRLGEI RDRSFEQQRY RASGLNLVTA AIVLWNTVYL ERAAHALRGN GHAVDDSLLQ YLSPLGWEHI NLTGDYLWRS SAKIGAGKFR PLRPLQPA |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| Tn3.1-KY749247.1 | Tn3.1 | transposon | 1019-5966 | + | 4948 |
| In_Tn4-KY749247.1 | In_Tn4 | integron | 9587-18970 | + | 9384 |
| IS1326-KY749247.1 | IS1326 | insertion sequence | 12265-14734 | + | 2470 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRL | Tn3.1 | 1019-1056 | + | 38 | GGGGTCTGAC GCTCAGTGGA ACGAAAACTC ACGTTAAG |
| IRR | Tn3.1 | 5929-5966 | - | 38 | GGGGTCTGAC GCTCAGTGGA ACGAAAACTC ACGTTAAG |
| IRt | In_Tn4 | 9587-9619 | + | 33 | TGTCATTTTC AGAAGACGAC TGCACCAGTT GAT |
| repeat t1 | In_Tn4 | 9595-9613 | + | 19 | TCAGAAGACG ACTGCACCA |
| repeat t2 | In_Tn4 | 9635-9653 | - | 19 | TCAGGAGCTG GCTGCACAA |
| repeat t3 | In_Tn4 | 9664-9683 | + | 20 | TCAGAAGTGA TCTGCACCAA |
| repeat t4 | In_Tn4 | 9696-9714 | + | 19 | TCAATACTCG TGTGCACCA |
| IRL | IS1326 | 12265-12290 | + | 26 | TGTTGAGTTG CATCTAAAAT TGACCC |
| IRR | IS1326 | 14709-14734 | - | 26 | TGTTGATTTG CACCCAAATT TGACCC |
| repeat i4 | In_Tn4 | 18851-18869 | - | 19 | TCAGCGGACG CAGGGAGGA |
| repeat i3 | In_Tn4 | 18879-18897 | - | 19 | TCAGGCAACG ACGGGCTGC |
| repeat i2 | In_Tn4 | 18921-18939 | - | 19 | TCAGAAGCCG ACTGCACTA |
| IRi | In_Tn4 | 18938-18970 | - | 33 | TGTCGTTTTC AGAAGACGGC TGCACTGAAC GTC |
| repeat i1 | In_Tn4 | 18944-18962 | - | 19 | TCAGAAGACG GCTGCACTG |
1. Cox KEL, Schildbach JF.
Sequence of the R1 plasmid and comparison to F and R100.. Plasmid. 2017 May;91:53-60. doi: 10.1016/j.plasmid.2017.03.007. Epub 2017 Mar 28. PubMed ID:
28359666
.