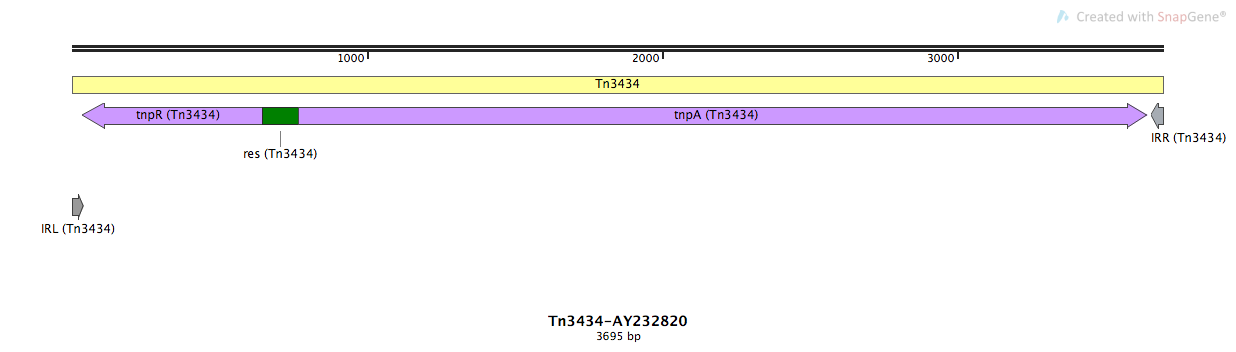
Mobile element type:
Transposon
Name:
Tn3434
Synonyms:
Accession:
Tn3434-AY232820
Family:
Tn3
Group:
Tn5393
First isolate:
Yes
Partial:
ND
Evidence of transposition:
Yes
Host Organism:
Paracoccus pantotrophus DSM11027
Date of Isolation:
2003
Country:
Poland
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-39 | + | 39 | GGGGTCTGAT GCCGGAGAGT GCAGAATCCC ACGCTAAGC |
| IRR | 3658-3695 | - | 38 | GGGGTCTGAT GCCGGAGAGT GCAGAATCCT ACGCTAAG |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res | Tn3434 | 646-765 | + | 120 | GTTTCACAAA CGTTCGTTTG AGGTGTTCTG GAAAGCGGCG CGCTTTCGGC CATCCGAACA ATTCCTTATT CAATATCTTA ATCATCAATA CGCAAACCAT ATGTAGAAAG CAAAAGAAAT |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| tnpR | Tn3434 | 34-645 | - | Accessory Gene | Resolvase |
| tnpA | Tn3434 | 766-3648 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn3434 | 203 | 34-645 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Function: | resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MLIGYARVSK ADGSQSLDLQ RDALIASGVE EDQIYSDLAS GKKDDRSGLE ACLKALREGD VLIVWKLDRL GRSLHHLVKT VSMLSERGVG LKVLTGQGAQ IDTTTAAGRL SFGIFAALAE FESELIRERT MAGLQAARAR GRKGGRKFAL TKAQVRMAQA AMASRDTSVS ELCKELGVKP VTLYRYVDPN GNLRDYGKRV LSA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn3434 | 960 | 766-3648 | + |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | identical totniA (Tn1721) | ||||||||||
| Protein Sequence: | MAHRTILTER QRSVLFDLPI DETAMLRHYI LADDDLEIIR ARRRPHNRFG FALQLCALRY PGRLLTPGEV IPMEITRFLA AQLGLKSDDL AGYASREETR HEHLAALRDL YGYKMFTGRG SRDLKAWLER TAETARSNED LARRFVEQCR ATQTILPGIT VIERLCADAL VAAERRIDAR IADRLDDEMC SRLDALLTET ADSSVTRFVW LRQFEVGQNS ADMNRLLDRL EFLQAMAVDR SILSGVPPHR IARLRRQGER YFAGDLRDIS GDRRLAILAV CALEWRSAIA DAVVESHDRI VGKTWREAKR VCDARADDAK AALKDTLQGF SNFGSALLEA YEDQASLMEA VQNAGGWSSL RGLVSTAAQL TDTLAADPLA HVVHGYHRFR RYAPRMLRAL DIQAAPVAEP LLAAADIVAG TETTTTRPLT FLRRASKWHR HLNHDDGNRL WEVAVLCHLR DAFRAGDIWL AHSRRYGDLK DALVPTEVAR ATPKLAMPFE PELWLADRKS RLADGLERLA RAARAGAIPG GSIEDGVLKI DRLTAAVPEE ADAMVLDLYN RLPEIRITDL LLEVDDEIGF TEAFTHLRTG VPCKDRVGML NVLLAEGLNL GLSKMAGATN THDYFQLSRL SRWHVESEAM ARALAMVIEG QSALPMARFW GAGQTASSDG QFFPTTRQGE AMNLINAKYG HEPGLKAYTH VSDQFGPFAT QTIPATVNEA PYILDGLLMT DAGQKIREQY ADTGGFTDHV FAVTALLGFQ FIPRIRDLPS KRLYLFNPAS CPKELKGLIG GKVREPVISS NWPDILRAAA TMVAGAMPPS QLLRKFAAYP RQHELAVALR EIGRIERTLF IIDWLLDADM QRRAQIGLNK GEAHHALKNA LRIGRQGEIR DRTAQGQHFR MAGLNLLAAI IIYWNTRHLG HAVDSRRSDG LDCSANLLAH ISPLGWAHIL LTGEYRWPKR |
||||||||||
1. Bartosik D, Sochacka M, Baj J.
Identification and characterization of transposable elements of Paracoccus pantotrophus.. J Bacteriol. 2003 Jul;185(13):3753-63. doi: 10.1128/JB.185.13.3753-3763.2003. PubMed ID:
12813068
.
2. Mikosa M, Sochacka-Pietal M, Baj J, Bartosik D. Identification of a transposable genomic island of Paracoccus pantotrophus DSM 11072 by its transposition to a novel entrapment vector pMMB2.. Microbiology (Reading). 2006 Apr;152(Pt 4):1063-1073. doi: 10.1099/mic.0.28603-0. PubMed ID: 16549670 .
2. Mikosa M, Sochacka-Pietal M, Baj J, Bartosik D. Identification of a transposable genomic island of Paracoccus pantotrophus DSM 11072 by its transposition to a novel entrapment vector pMMB2.. Microbiology (Reading). 2006 Apr;152(Pt 4):1063-1073. doi: 10.1099/mic.0.28603-0. PubMed ID: 16549670 .