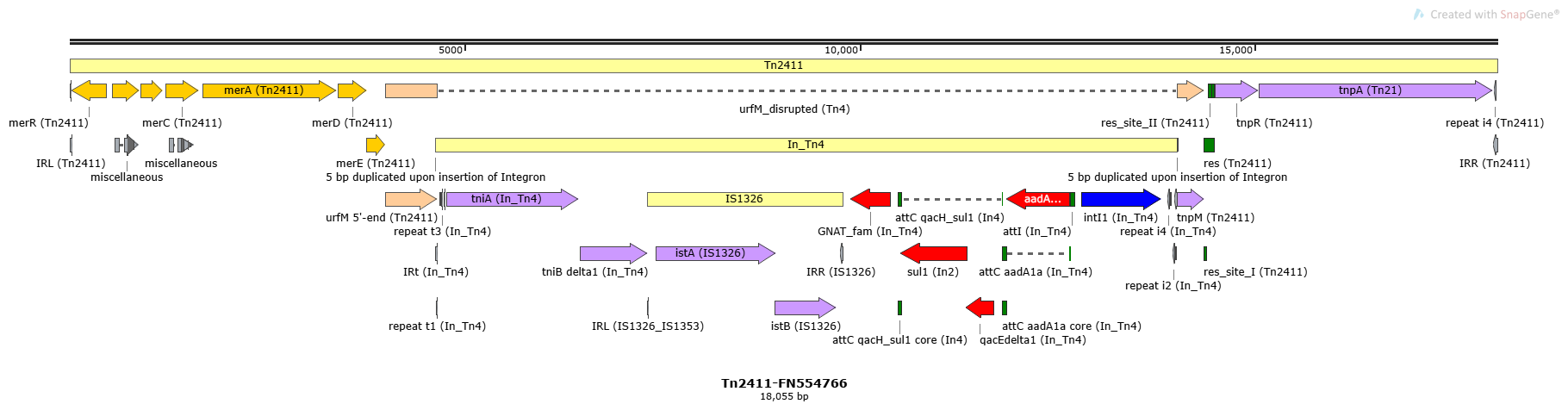
Mobile element type:
Transposon
Name:
Tn2411
Synonyms:
Accession:
Tn2411-FN554766
Family:
Tn3
Group:
Tn21
First isolate:
Yes
Partial:
ND
Evidence of transposition:
ND
Host Organism:
Escherichia coli 042
Date of Isolation:
2010
Country:
Molecular Source:
chromosome
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-37 | + | 37 | GGGGCACCTC AGAAAACGGA AAATAAAGCA CGCTAAG |
| repeat i4 | 9-27 | + | 19 | TCAGAAAACG GAAAATAAA |
| IRR | 18015-18055 | - | 41 | GGGGTCGTCT CAGAAAACGG AAAATAAAGC ACGCTAAGCC G |
| repeat i4 | 18028-18046 | - | 19 | TCAGAAAACG GAAAATAAA |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| attC aadA1a core | In_Tn4 | 11806-11859 | + | 54 | CGCTTGAGTT AAGCCGCGCC GCGAAGCGGC GTCGGCTTGA ACGAATTGTT AGAC |
| attI | In_Tn4 | 12662-12717 | + | 56 | CTTTGTTTTA GGGCGACTGC CCTGCTGCGT AACATCGTTG CTGCTCCATA ACATCA |
| res | Tn2411 | 14352-14482 | + | 131 | GCCGCCGTCA GGTTGAGGCA TACCCTAACC TGATGTCAGA TGCCATGTGT AAATTGCGTC AGGATAGGAT TGAATTTTGA ATTTATTGAC ATATCTCGTT GAAGGTCATA GAGTCTTCCC TGACATTTTG C |
| res_site_I | Tn2411 | 14352-14390 | + | 39 | GCCGCCGTCA GGTTGAGGCA TACCCTAACC TGATGTCAG |
| res_site_II | Tn2411 | 14404-14447 | + | 44 | ATTGCGTCAG GATAGGATTG AATTTTGAAT TTATTGACAT ATCT |
| res_site_III | Tn2411 | 14451-14482 | + | 32 | TGAAGGTCAT AGAGTCTTCC CTGACATTTT GC |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| merR | Tn2411 | 33-467 | - | Passenger Gene | Heavy Metal Resistance |
| merT | Tn2411 | 539-889 | + | Passenger Gene | Heavy Metal Resistance |
| merP | Tn2411 | 903-1178 | + | Passenger Gene | Heavy Metal Resistance |
| merC | Tn2411 | 1214-1636 | + | Passenger Gene | Heavy Metal Resistance |
| merA | Tn2411 | 1688-3382 | + | Passenger Gene | Heavy Metal Resistance |
| merD | Tn2411 | 3400-3762 | + | Passenger Gene | Heavy Metal Resistance |
| merE | Tn2411 | 3759-3995 | + | Passenger Gene | Heavy Metal Resistance |
| urfM 5'-end | Tn2411 | 3992-4662 | + | Passenger Gene | Other |
| tniA | In_Tn4 | 4774-6453 | + | Transposase | |
| tniB delta1 | In_Tn4 | 6456-7316 | + | Accessory Gene | |
| istA | IS1326 | 7418-8941 | + | Transposase | None |
| istB | IS1326 | 8928-9713 | + | Accessory Gene | ATPase Transposition Helper |
| GNAT_fam | In_Tn4 | 9889-10389 | - | Passenger Gene | Antibiotic Resistance |
| qacEdelta1 (ARO:3005010) | In_Tn4 | 11350-11697 | - | Passenger Gene | Antibiotic Resistance |
| aadA (ARO:3002601) | In_Tn4 | 11861-12652 | - | Passenger Gene | Antibiotic Resistance |
| intI1 | In_Tn4 | 12801-13814 | + | Integron Integrase | Class 1 |
| tnpM | Tn2411 | 14017-14367 | + | Accessory Gene | Inhibitor |
| tnpR | Tn2411 | 14493-15053 | + | Accessory Gene | Resolvase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merR | MerR | Tn2411 | 144 | 33-467 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | activator-repressor of mer operon | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MENNLENLTI GVFAKAAGVN VETIRFYQRK GLLREPDKPY GSIRRYGEAD VVRVKFVKSA QRLGFSLDEI AELLRLDDGT HCEEASSLAE HKLKDVREKM ADLARMETVL SELVCACHAR KGNVSCPLIA SLQGEAGLAR SAMP |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merT | MerT | Tn2411 | 116 | 539-889 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | cytosolic mercuric ion transport protein | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MSEPQNGRGA LFAGGLAAIL ASTCCLGPLV LVALGFSGAW IGNLTVLEPY RPLFIGAALV ALFFAWKRIY RPVQACKPGE VCAIPQVRAT YKLIFWIVAV LVLVALGFPY VVPFFY |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merP | MerP | Tn2411 | 91 | 903-1178 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercury transport | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MKKLFASLAL AAAVAPVWAA TQTVTLAVPG MTCAACPITV KKALSKVEGV SKVDVGFEKR EAVVTFDDTK ASVQKLTKAT ADAGYPSSVK Q |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merC | MerC | Tn2411 | 140 | 1214-1636 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | transmembrane protein mercury transport | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MGLMTRIADK TGALGSVVSA MGCAACFPAL ASFGAAIGLG FLSQYEGLFI SRLLPLFAAL AFLANALGWF SHRQWLRSLL GMIGPAIVFA ATVWLLGNWW TANLMYVGLA LMIGVSIWDF VSPAHRRCGP DGCELPAKRL |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merA | MerA | Tn2411 | 564 | 1688-3382 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercuric ion reductase | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MSTLKITGMT CDSCAVHVKD ALEKVPGVQS ADVSYAKGSA KLAIEVGTSP DALTAAVAGL GYRATLADAP SVSTPGGLLD KMRDLLGRND KTGSSGALHI AVIGSGGAAM AAALKAVEQG ARVTLIERGT IGGTCVNVGC VPSKIMIRAA HIAHLRRESP FDGGIAATTP TIQRTALLAQ QQARVDELRH AKYEGILEGN PAITVLHGSA RFKDNRNLIV QLNDGGERVV AFDRCLIATG ASPAVPPIPG LKDTPYWTST EALVSETIPK RLAVIGSSVV ALELAQAFAR LGAKVTILAR STLFFREDPA IGEAVTAAFR MEGIEVREHT QASQVAYING EGDGEFVLTT AHGELRADKL LVATGRAPNT RKLALDATGV TLTPQGAIVI DPGMRTSVEH IYAAGDCTDQ PQFVYVAAAA GTRAAINMTG GDAALNLTAM PAVVFTDPQV ATVGYSEAEA HHDGIKTDSR TLTLDNVPRA LANFDTRGFI KLVVEEGSGR LIGVQAVAPE AGELIQTAAL AIRNRMTVQE LADQLFPYLT MVEGLKLAAQ TFNKDVKQLS CCAG |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merD | MerD | Tn2411 | 120 | 3400-3762 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | secondary regulatory protein | ||||||||||
| Target: | Mercury | ||||||||||
| Protein Sequence: | MSAYTVSQLA HNAGVSVHIV RDYLVRGLLR PVACTTGGYG VFDDAALQRL CFVRAAFEAG IGLDALARLC RALDAADGAQ AAAQLAVLRQ LVERRRAALA HLDAQLASMP AERAHEEALP |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | merE | MerE | Tn2411 | 78 | 3759-3995 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Heavy Metal Resistance | ||||||||||
| Function: | mercury transport | ||||||||||
| Target: | Mercury | ||||||||||
| Comment: | similar to urf-1 in pKLH2 (GenBank AF213017), pKLH272 (Genbank Y08992), pMER610 (GenBank Y08993), pKLH210 (GenBank Y10102), Tn5036 (Genbank Y09025), orf1 in Tn501 (GenBank Z00027), and urf-1 in Tn5041 (GenBank X98999) | ||||||||||
| Protein Sequence: | MNAPDKLPPE TRQPVSGYLW GALAVLTCPC HLPILAAVLA GTTAGAFLGE HWGVAALALT GLFVLAVTRL LRAFRGGS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | urfM 5'-end | N/A | Tn2411 | 223 | 3992-4662 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Comment: | urfMORF interrupted by insertion of In2 | ||||||||||
| Protein Sequence: | MTSSQPAGWT AAELAQAAAR GQLDLHYQPL VDLRDHRIAG AEALMRWRHP RLGLLPPGQF LPLAESFGLM PEIGAWVLGE ACRQMHKWQG PAWQPFRLAI NVSASQVGPT FDDEVKRVLA DMALPAELLE IELTESVAFG NPALFASFDA LRAIGVRFAA DDFGTGYSCL QHLKCCPITT LKIDQSFVAR LPDDARDQTI VRAVIQLAHG LGMDVIFRRR LHQ |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniA | TniA | In_Tn4 | 559 | 4774-6453 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | can be extended upstream by 12 amino acids| identical to tniA (Tn1721 and In2)| 25% amino acid sequence identity to TnsB from Tn7 | ||||||||||
| Protein Sequence: | MATDTPRIPE QGVATLPDEA WERARRRAEI ISPLAQSETV GHEAADMAAQ ALGLSRRQVY VLIRRARQGS GLVTDLVPGQ SGGGKGKGRL PEPVERVIHE LLQKRFLTKQ KRSLAAFHRE VTQVCKAQKL RVPARNTVAL RIASLDPRKV IRRREGQDAA RDLQGVGGEP PAVTAPLEQV QIDHTVIDLI VVDDRDRQPI GRPYLTLAID VFTRCVLGMV VTLEAPSAVS VGLCLVHVAC DKRPWLEGLN VEMDWQMSGK PLLLYLDNAA EFKSEALRRG CEQHGIRLDY RPLGQPHYGG IVERIIGTAM QMIHDELPGT TFSNPDQRGD YDSENKAALT LRELERWLTL AVGTYHGSVH NGLLQPPAAR WAEAVARVGV PAVVTRATSF LVDFLPILRR TLTRTGFVID HIHYYADALK PWIARRERWP SFLIRRDPRD ISRIWVLEPE GQHYLEIPYR TLSHPAVTLW EQRQALAKLR QQGREQVDES ALFRMIGQMR EIVTSAQKAT RKARRDADRR QHLKTSARPD KPVPPDTDIA DPQADNLPPA KPFDQIEEW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniB delta1 | TniB delta1 | In_Tn4 | 286 | 6456-7316 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Function: | probable ATP-binding protein. | ||||||||||
| Comment: | probably truncated by insertion of IS1326::IS1353 | ||||||||||
| Protein Sequence: | MDEYPIIDLS HLLPAAQGLA RLPADERIQR LRADRWIGYP RAVEALNRLE ALYAWPNKQR MPNLLLVGPT NNGKSMIVEK FRRTHPASSD ADQEHIPVLV VQMPSEPSVI RFYVALLAAM GAPLRPRPRL PEMEQLALAL LRKVGVRMLV IDELHNVLAG NSVNRREFLN LLRFLGNELR IPLVGVGTRD AYLAIRSDDQ LENRFEPMML PVWEANDDCC SLLASFAASL PLRRPSPIAT LDMARYLLTR SEGTIGELAH LLMAAAIVAV ESGEEAINHR TLSMAC |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | istA | IstA | IS1326 | 507 | 7418-8941 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MINVAILSAI RRWHFRDGAS IREIARRSGL SRNTVRKYLQ SKVVEPQYPA RDSVGKLSPF EPKLRQWLST EHKKTKKLRR NLRSMYRDLV ALGFTGSYDR VCAFARQWKD SEQFKAQTSG KGCFIPLRFA CGEAFQFDWS EDFARIAGKQ VKLQIAQFKL AHSRAFVLRA YYQQKHEMLF DAHWHAFQIF GGIPKRGIYD NMKTAVDSVG RGKERRVNQR FTAMVSHYLF DAQFCNPASG WEKGQIEKNV QDSRQRLWQG APDFQSLADL NVWLEHRCKA LWSELRHPEL DQTVQEAFAD EQGELMALPN AFDAFVEQTK RVTSTCLVHH EGNRYSVPAS YANRAISLRI YADKLVMAAE GQHIAEHPRL FGSGHARRGH TQYDWHHYLS VLQKKPGALR NGAPFAELPP AFKKLQSILL QRPGGDRDMV EILALVLHHD EGAVLSAVEL ALECGKPSKE HVLNLLGRLT EEPPPKPIPI PKGLRLTLEP QANVNRYDSL RRAHDAA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | istB | IstB | IS1326 | 261 | 8928-9713 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | ATPase Transposition Helper | ||||||||||
| Function: | stimulates transposition | ||||||||||
| Protein Sequence: | MMQHEGHVRI LKSLKLFGMA HAIEELGNQN SPAFNQALPM LDSLIKAEVA EREVRSVNYQ LRVAKFPVYR DLVGFDFSQS LVNEATVKQL HRCDFMEQAQ NVVLIGGPGT GKTHLATAIG TQAVMHLNRR VRFFSTVDLV NALEQEKSSG RQGQIANRLL YADLVILDEL GYLPFSQTGG ALLFHLLSKL YEKTSVILTT NLSFSEWSRV FGDEKMTTAL LDRLTHHCHI LETGNESYRF KHSSTQNKQE EKQTRKLKIE T |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | GNAT_fam | GNAT_fam | In_Tn4 | 166 | 9889-10389 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Sequence Family: | Acetyltransf_1 (Pfam:PF00583) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | putative acetyltransferase ADU64769.1 | ||||||||||
| Protein Sequence: | MDSEEPPNVR VACSGDIDEV VRLMHDAAAW MSAKGTPAWD VARIDRTFAE TFVLRSELLV ASCSDGIVGC CTLSAEDPEF WPDALKGEAA YLHKLAVRRT HAGRGVSSAL IEACRHAART QGCAKLRLDC HPNLRGLYER LGFTHVDTFN PGWDPTFIAE RLELEI |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | qacEdelta1 (ARO:3005010) | QacEdelta1 | In_Tn4 | 115 | 11350-11697 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic efflux (ARO:0010000) | ||||||||||
| Sequence Family: | major facilitator superfamily (MFS) antibiotic efflux pump (ARO:0010002) | ||||||||||
| Target: | disinfecting agents and antiseptics(ARO:3005386) | ||||||||||
| Comment: | subunit of the qac multidrug efflux pump||perfect match to reference sequence for ARO:3005010 (bitscore:219) | ||||||||||
| Protein Sequence: | MKGWLFLVIA IVGEVIATSA LKSSEGFTKL APSAVVIIGY GIAFYFLSLV LKSIPVGVAY AVWSGLGVVI ITAIAWLLHG QKLDAWGFVG MGLIIAAFLL ARSPSWKSLR RPTPW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | aadA (ARO:3002601) | AadA | In_Tn4 | 263 | 11861-12652 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | ANT(3'')(ARO:3004275) | ||||||||||
| Transposase Chemistry: | aminoglycoside nucleotidyltransferase | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | perfect match to reference sequence for ARO:3002601||Synonyms: aadA1-pm, aadA, aadA1, aad(3'')(9) | ||||||||||
| Protein Sequence: | MREAVIAEVS TQLSEVVGVI ERHLEPTLLA VHLYGSAVDG GLKPHSDIDL LVTVTVRLDE TTRRALINDL LETSASPGES EILRAVEVTI VVHDDIIPWR YPAKRELQFG EWQRNDILAG IFEPATIDID LAILLTKARE HSVALVGPAA EELFDPVPEQ DLFEALNETL TLWNSPPDWA GDERNVVLTL SRIWYSAVTG KIAPKDVAAD WAMERLPAQY QPVILEARQA YLGQEEDRLA SRADQLEEFV HYVKGEITKV VGK |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | intI1 | IntI1 | In_Tn4 | 337 | 12801-13814 | + |
|---|
| Class: | Integron Integrase | ||||||||||
| Subclass: | Class 1 | ||||||||||
| Sequence Family: | Class 1 Integron Tyrosine Integrase | ||||||||||
| Transposase Chemistry: | Tyrosine | ||||||||||
| Protein Sequence: | MKTATAPLPP LRSVKVLDQL RERIRYLHYS LRTEQAYVHW VRAFIRFHGV RHPATLGSSE VEAFLSWLAN ERKVSVSTHR QALAALLFFY GKVLCTDLPW LQEIGRPRPS RRLPVVLTPD EVVRILGFLE GEHRLFAQLL YGTGMRISEG LQLRVKDLDF DHGTIIVREG KGSKDRALML PESLAPSLRE QLSRARAWWL KDQAEGRSGV ALPDALERKY PRAGHSWPWF WVFAQHTHST DPRSGVVRRH HMYDQTFQRA FKRAVEQAGI TKPATPHTLR HSFATALLRS GYDIRTVQDL LGHSDVSTTM IYTHVLKVGG AGVRSPLDAL PPLTSER |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpM | TnpM | Tn2411 | 116 | 14017-14367 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Inhibitor | ||||||||||
| Function: | transposition regulator||reported to enhance Tn21 transposition and suppress resolution of cointegrate replicons in vivo | ||||||||||
| Comment: | 3'-end of urfM ORF, which is interrupted by insertion of In2||inhibits tranposition probably by inhibiting resolution | ||||||||||
| Protein Sequence: | MEVVAEGVET PDCLAWLRQA GCDTVQGFLF ARPMPAAAFV GFVNQWRNTT MNANEPSTSC CVCCKEIPLD AAFTPEGAEY VEHFCGLECY QRFQARASTA TETSVKPDAC DSPPSG |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn2411 | 186 | 14493-15053 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Function: | resolvase||serine site-specific recombinase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Comment: | identical to tnpR (TnAs3 ) | ||||||||||
| Protein Sequence: | MTGQRIGYIR VSTFDQNPER QLEGVKVDRA FSDKASGKDV KRPQLEALIS FARTGDTVVV HSMDRLARNL DDLRRIVQTL TQRGVHIEFV KEHLSFTGED SPMANLMLSV MGAFAEFERA LIRERQREGI ALAKQRGAYR GRKKSLSSER IAELRQRVEA GEQKTKLARE FGISRETLYQ YLRTDQ |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| In_Tn4-KY749247.1 | In_Tn4 | integron | 4633-14016 | + | 9384 |
| IS1326-KY749247.1 | IS1326 | insertion sequence | 7311-9780 | + | 2470 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRt | In_Tn4 | 4633-4665 | + | 33 | TGTCATTTTC AGAAGACGAC TGCACCAGTT GAT |
| repeat t1 | In_Tn4 | 4641-4659 | + | 19 | TCAGAAGACG ACTGCACCA |
| repeat t2 | In_Tn4 | 4681-4699 | - | 19 | TCAGGAGCTG GCTGCACAA |
| repeat t3 | In_Tn4 | 4710-4729 | + | 20 | TCAGAAGTGA TCTGCACCAA |
| repeat t4 | In_Tn4 | 4742-4760 | + | 19 | TCAATACTCG TGTGCACCA |
| IRR | IS1326 | 9755-9780 | - | 26 | TGTTGATTTG CACCCAAATT TGACCC |
| repeat i4 | In_Tn4 | 13897-13915 | - | 19 | TCAGCGGACG CAGGGAGGA |
| repeat i3 | In_Tn4 | 13925-13943 | - | 19 | TCAGGCAACG ACGGGCTGC |
| repeat i2 | In_Tn4 | 13967-13985 | - | 19 | TCAGAAGCCG ACTGCACTA |
| IRi | In_Tn4 | 13984-14016 | - | 33 | TGTCGTTTTC AGAAGACGGC TGCACTGAAC GTC |
| repeat i1 | In_Tn4 | 13990-14008 | - | 19 | TCAGAAGACG GCTGCACTG |
1. Chaudhuri RR, Sebaihia M, Hobman JL, Webber MA, Leyton DL, Goldberg MD, Cunningham AF, Scott-Tucker A, Ferguson PR, Thomas CM, Frankel G, Tang CM, Dudley EG, Roberts IS, Rasko DA, Pallen MJ, Parkhill J, Nataro JP, Thomson NR, Henderson IR.
Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042.. PLoS One. 2010 Jan 20;5(1):e8801. doi: 10.1371/journal.pone.0008801. PubMed ID:
20098708
.