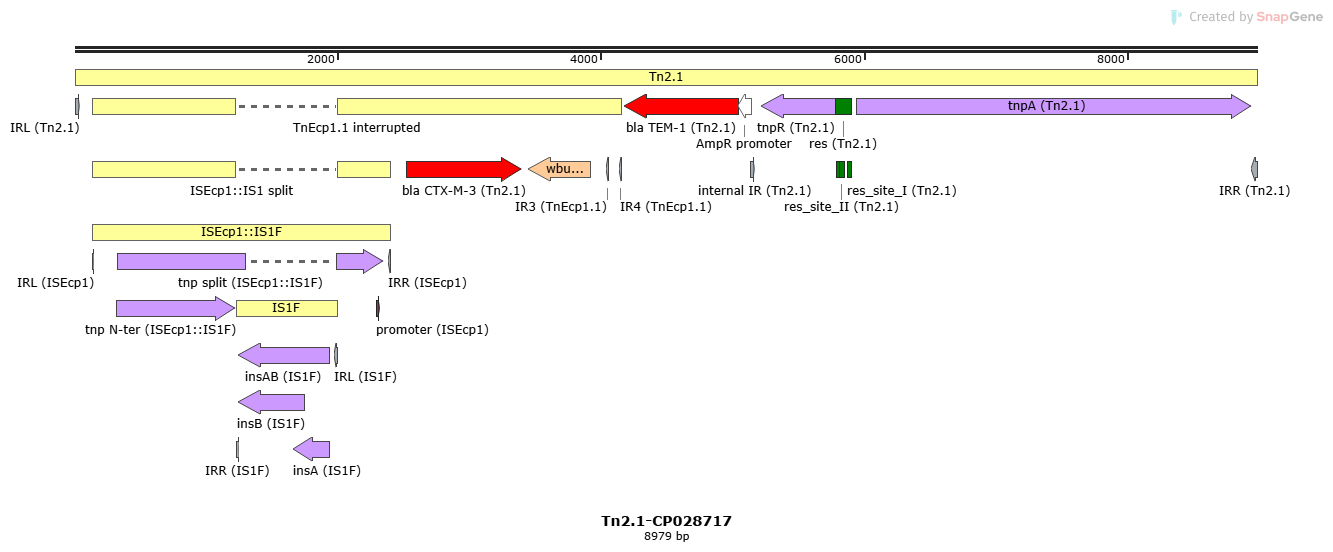
Mobile element type:
Transposon
Name:
Tn2.1
Synonyms:
Accession:
Tn2.1-CP028717
Family:
Tn3
Group:
Tn3
First isolate:
Yes
Partial:
ND
Evidence of transposition:
Yes
Host Organism:
Klebsiella pneumoniae SCM96
Date of Isolation:
2018
Country:
China
Molecular Source:
plasmid pSCM96-1
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-38 | + | 38 | GGGGTCTGAC GCTCAGTGGA ACGAAAACTC ACGTTAAG |
| internal IR | 5132-5169 | + | 38 | GGGGTTCCGC GCACATTTCC CCGAAAAGTG CCACCTGA |
| IRR | 8942-8979 | - | 38 | GGGGTCTGAC GCTCAGTGGA ACGAAAACTC ACGTTAAG |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res | Tn2.1 | 5780-5900 | + | 121 | AAATGTACCT TAAATCGAAT ATCGGACAAC TCATGTCTAT TATTACAAAT TTACGATTTA ATAGACATAT TAATGTAACA GTTTTACGAT GTCCGATAAT TTATAACATT TCGTACGGTT G |
| res_site_III | Tn2.1 | 5783-5805 | + | 23 | TGTACCTTAA ATCGAATATC GGA |
| res_site_II | Tn2.1 | 5813-5846 | + | 34 | TGTCTATTAT TACAAATTTA CGATTTAATA GACA |
| res_site_I | Tn2.1 | 5869-5897 | + | 29 | TGTCCGATAA TTTATAACAT TTCGTACGG |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| tnp N-ter | ISEcp1::IS1F | 316-1221 | + | Transposase | |
| insAB | IS1F | 1237-1934 | - | Transposase | |
| insB | IS1F | 1237-1740 | - | Transposase | |
| insA | IS1F | 1659-1934 | - | Accessory Gene | |
| bla CTX-M-3 (ARO:3001866) | Tn2.1 | 2520-3395 | + | Passenger Gene | Antibiotic Resistance |
| wbuC family | Tn2.1 | 3442-3918 | - | Passenger Gene | Other |
| bla TEM-1 (ARO:3000873) | Tn2.1 | 4177-5037 | - | Passenger Gene | Antibiotic Resistance |
| tnpR | Tn2.1 | 5220-5777 | - | Accessory Gene | Resolvase |
| tnpA | Tn2.1 | 5941-8946 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnp N-ter | Tnp N-ter | ISEcp1::IS1F | 302 | 316-1221 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | tnp split by insertion of ISF1 | ||||||||||
| Protein Sequence: | MINKIDFKAK NLTSNAGLFL LLENAKSNGI FDFIENDLVF DNDSTNKIKM NHIKTMLCGH FIGIDKLERL KLLQNDPLVN EFDISVKEPE TVSRFLGNFN FKTTQMFRDI NFKVFKKLLT KSKLTSITID IDSSVINVEG HQEGASKGYN PKKLGNRCYN IQFAFCDELK AYVTGFVRSG NTYTANGAAE MIKEIVANIK SDDLEILFRM DSGYFDEKII ETIESLGCKY LIKAKSYSTL TSQATNSSIV FVKGEEGRET TELYTKLVKW EKDRRFVVSR VLKPEKERAQ LSLLEGSEYD YF |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | insAB | InsAB | IS1F | 232 | 1237-1934 | - |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | fusion protein from -1 programmed frameshifting between insA and insB | ||||||||||
| Protein Sequence: | VASISISCPS CSATEGVVRN GKSTAGHQRY LCSHCRKTWQ LQFTYTASQP GTHQKIIDMA MNGVGCRATA RIMGVGLNTI LRHLKNSGRS R*PRAYSRAV M*LSALKWTN SGATSVLNHV SAGCFTRMTG YGGRLWRTSS VNALWPHWSV F*ACCRPLRS WYG*RMAGRC MNPA*RESCT *SASVTLSAL SDIT*I*DNI WQGWDGSHCR SQNRWSCMTK SSGII*T*NT IS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | insB | InsB | IS1F | 167 | 1237-1740 | - |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | downstream catalytic domain | ||||||||||
| Protein Sequence: | MPGNRPHYGR WPQHDFTSLK KLRPQSVTSR IQPGSDVIVC AEMDEQWGYV GAKSRQRWLF YAYDRIRRTV VAHVFGERTL ATLERLLSLL SAFEVVVWMT DGWPLYESRL KGKLHVISKR YTQRIERHNL NLRQHLARLG RKSLSFSKSV ELHDKVIGHY LNIKHYQ |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | insA | InsA | IS1F | 91 | 1659-1934 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Comment: | regulatory protein | ||||||||||
| Protein Sequence: | VASISISCPS CSATEGVVRN GKSTAGHQRY LCSHCRKTWQ LQFTYTASQP GTHQKIIDMA MNGVGCRATA RIMGVGLNTI LRHLKNSGRS R |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | bla CTX-M-3 (ARO:3001866) | Bla CTX-M-3 | Tn2.1 | 291 | 2520-3395 | + |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | CTX-M beta-lactamase (ARO:3000016) | ||||||||||
| Target: | cephalosporin (ARO:0000032)||penam (ARO:3000008)||cephamycin (ARO:0000044) | ||||||||||
| Comment: | strict match to reference sequence for ARO:3001866 (bitscore: 594)||Synonyms: | ||||||||||
| Protein Sequence: | MVKKSLRQFT LMATATVTLL LGSVPLYAQT ADVQQKLAEL ERQSGGRLGV ALINTADNSQ ILYRADERFA MCSTSKVMAA AAVLKKSESE PNLLNQRVEI KKSDLVNYNP IAEKHVNGTM SLAELSAAAL QYSDNVAMNK LIAHVGGPAS VTAFARQLGD ETFRLDRTEP TLNTAIPGDP RDTTSPRAMA QTLRNLTLGK ALGDSQRAQL VTWMKGNTTG AASIQAGLPA SWVVGDKTGS GDYGTTNDIA VIWPKDRAPL ILVTYFTQPQ PKAESRRDVL ASAAKIVTDG L |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | wbuC family | WbuC family | Tn2.1 | 158 | 3442-3918 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Comment: | cupin fold metalloprotein | ||||||||||
| Protein Sequence: | MKQITMSDMQ QQSAAAVQSP RLRAHRNFHP ELSDSVQRLA IAMEPGTYVR PHRHPHTFEL LLPLRGRFVV LNFDDRGTVT HRAILGETCT VLEMAAGTWH AVLSLDTGGI IFEVKHGGYQ PVAADDYAHW APAEGEPGTT ELMAWYAQAQ VGDSTFAV |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | bla TEM-1 (ARO:3000873) | Bla TEM-1 | Tn2.1 | 286 | 4177-5037 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | TEM beta-lactamase (ARO:3000014) | ||||||||||
| Target: | penam (ARO:3000008)||monobactam (ARO:0000004)||cephalosporin (ARO:0000032)||penem (ARO:3003706) | ||||||||||
| Comment: | 100 % identity with reference sequence ARO:3000873 (bitscore=591) | ||||||||||
| Protein Sequence: | MSIQHFRVAL IPFFAAFCLP VFAHPETLVK VKDAEDQLGA RVGYIELDLN SGKILESFRP EERFPMMSTF KVLLCGAVLS RVDAGQEQLG RRIHYSQNDL VEYSPVTEKH LTDGMTVREL CSAAITMSDN TAANLLLTTI GGPKELTAFL HNMGDHVTRL DRWEPELNEA IPNDERDTTM PAAMATTLRK LLTGELLTLA SRQQLIDWME ADKVAGPLLR SALPAGWFIA DKSGAGERGS RGIIAALGPD GKPSRIVVIY TTGSQATMDE RNRQIAEIGA SLIKHW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn2.1 | 185 | 5220-5777 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Function: | resolvase||serine site-specific recombinase (GO:0000150) | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MRLFGYARVS TSQQSLDLQV RALKDAGVKA NRIFTDKASG SSTDREGLDL LRMKVEEGDV ILVKKLDRLG RDTADMIQLI KEFDAQGVAV RFIDDGISTD GDMGQMVVTI LSAVAQAERR RILERTNEGR QEAKLKGIKF GRRRTVDRNV VLTLHQKGTG ATEIAHQLSI ARSTVYKILE DERAS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn2.1 | 1001 | 5941-8946 | + |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase activity (GO:0004803) | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | identical to tnpA (Tn1) | ||||||||||
| Protein Sequence: | MPVDFLTTEQ TESYGRFTGE PDELQLARYF HLDEADKEFI GKSRGDHNRL GIALQIGCVR FLGTFLTDMN HIPSGVRHFT ARQLGIRDIT VLAEYGQREN TRREHAALIR QHYQYREFAW PWTFRLTRLL YTRSWISNER PGLLFDLATG WLMQHRIILP GATTLTRLIS EVREKATLRL WNKLALIPSA EQRSQLEMLL GPTDCSRLSL LESLKKGPVT ISGPAFNEAI ERWKTLNDFG LHAENLSTLP AVRLKNLARY AGMTSVFNIA RMSPQKRMAV LVAFVLAWET LALDDALDVL DAMLAVIIRD ARKIGQKKRL RSLKDLDKSA LALASACSYL LKEETPDESI RAEVFSYIPR QKLAEIITLV REIARPSDDN FHEEMVEQYG RVRRFLPHLL NTVKFSSAPA GVTTLNACDY LSREFSSRRQ FFDDAPTEII SRSWKRLVIN KEKHITRRGY TLCFLSKLQD SLRRRDVYVT GSNRWGDPRA RLLQGADWQA NRIKVYRSLG HPTDPQEAIK SLGHQLDSRY RQVAARLCEN EAVELDVSGP KPRLTISPLA SLDEPDSLKR LSKMISDLLP PVDLTELLLE INAHTGFADE FFHASEASAR VDDLPVSISA VLMAEACNIG LEPLIRSNVP ALTRHRLNWT KANYLRAETI TSANARLVDF QATLPLAQIW GGGEVASADG MRFVTPVRTI NAGPNRKYFG NNRGITWYNF VSDQYSGFHG IVIPGTLRDS IFVLEGLLEQ ETGLNPTEIM TDTAGASELV FGLFWLLGYQ FSPRLADAGA SVFWRMDHDA DYGVLNDIAR GQSDPRKIVL QWDEMIRTAG SLKLGKVQAS VLVRSLLKSE RPSGLTQAII EVGRINKTLY LLNYIDDEDY RRRILTQLNR GESRHAVARA ICHGQKGEIR KRYTDGQEDQ LGALGLVTNA VVLWNTIYMQ AALDHLRAQG ETLNDEDIAR LSPLCHGHIN MLGHYSFTLA ELVTKGHLRP LKEASEAENV A |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| TnEcp1.1 interrupted | TnEcp1.1 interrupted | transposon | 130-1221 , 1990-4153 | + | 3256 |
| ISEcp1::IS1 split-Tn2.1-CP028717 | ISEcp1 split | insertion sequence | 130-1221 , 1990-2392 | + | 1495 |
| ISEcp1_IS1F-CP028717 | ISEcp1::IS1F | insertion sequence | 130-2392 | + | 2263 |
| IS1F-X52538 | IS1F | insertion sequence | 1222-1989 | + | 768 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRR | IS1F | 1222-1244 | + | 23 | GGTAATGACT CCAACTTACT GAT |
| IRL | IS1F | 1967-1989 | - | 23 | GGTAATGGTG CCAACTTACT GAT |