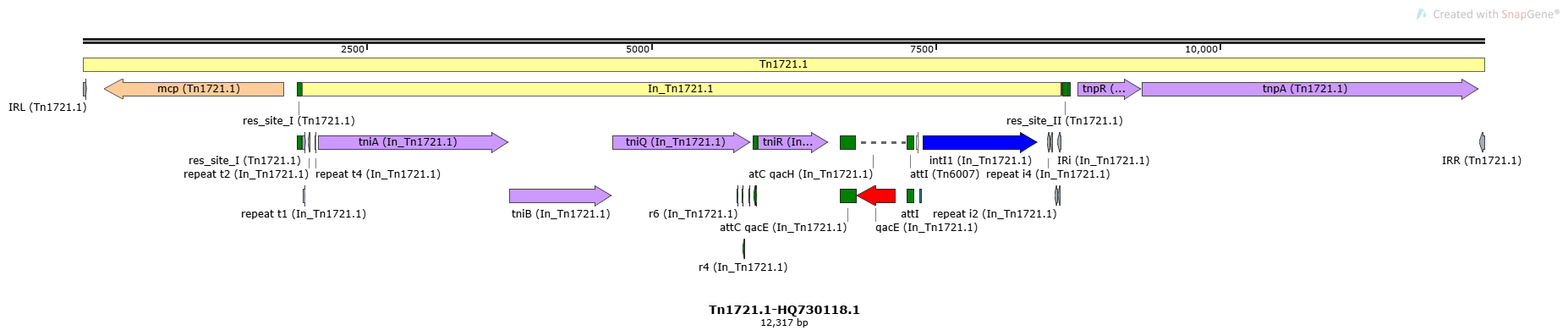
Mobile element type:
Transposon
Name:
Tn1721.1
Synonyms:
Accession:
Tn1721.1-HQ730118.1
Family:
Tn3
Group:
Tn21
First isolate:
Yes
Partial:
ND
Evidence of transposition:
ND
Host Organism:
Klebsiella pneumoniae WM2b02
Date of Isolation:
2011
Country:
Australia
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-38 | + | 38 | GGGGGAACCG CAGAATTCGG AAAAAATCGT ACGCTAAG |
| IRR | 12281-12317 | - | 37 | GGGGAGCCCG CAGAATTCGG AAAAAATCGT ACGCTAA |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| res_site_I | Tn1721.1 | 1886-1923 | + | 38 | GCGCATGTCA ATCTAGGCTA TACCCTAACT TGATGTCA |
| res_site_I | Tn1721.1 | 1886-1923 | + | 38 | GCGCATGTCA ATCTAGGCTA TACCCTAACT TGATGTCA |
| r6 | In_Tn1721.1 | 5755-5768 | + | 14 | TTGGAACGGT TGCG |
| r5 | In_Tn1721.1 | 5801-5814 | + | 14 | AATTTCTGTC ACAT |
| r4 | In_Tn1721.1 | 5806-5819 | - | 14 | AGGTAATGTG ACAG |
| r3 | In_Tn1721.1 | 5857-5870 | + | 14 | GGGTTAAGTG ACAA |
| res | In_Tn1721.1 | 5898-5932 | + | 35 | ACACTGTCAC ATAATCGAAC GTATACGTGA CGGGT |
| r2 | In_Tn1721.1 | 5901-5914 | - | 14 | CGATTATGTG ACAG |
| r1 | In_Tn1721.1 | 5917-5930 | + | 14 | CGTATACGTG ACGG |
| attC qacE | In_Tn1721.1 | 6659-6799 | + | 141 | GTTCAACGCC GAGTTCAGCG GCAGTTTTTA AGTTGTGATT TTATGGAATA CTTTTGCGCA GCAAAACCAT AAAGCCGCGA CTTAAAAACT GTCCAGCGCA GGCACGAAGT GCTGGAGCGG TGCTGCAACG ACTTGTTAGA T |
| res_site_II | Tn1721.1 | 8615-8658 | + | 44 | GCTTCGTCAG AATAGAGTCT GCTTTCCCAT TTTTTGACAC ATGC |
| res_site_III | Tn1721.1 | 8662-8687 | + | 26 | CGAAGGTTAT AGATTTCAGC CTGACA |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| mcp | Tn1721.1 | 190-1767 | - | Passenger Gene | Other |
| tniA | In_Tn1721.1 | 2073-3752 | + | Transposase | |
| tniB | In_Tn1721.1 | 3755-4663 | + | Accessory Gene | |
| tniQ | In_Tn1721.1 | 4660-5877 | + | Accessory Gene | Target Site Selection |
| tniR | In_Tn1721.1 | 5939-6562 | + | Accessory Gene | Resolvase |
| qacE (ARO:3005009) | In_Tn1721.1 | 6811-7143 | - | Passenger Gene | Antibiotic Resistance |
| intI1 | In_Tn1721.1 | 7388-8401 | + | Integron Integrase | Class 1 |
| tnpR | Tn1721.1 | 8754-9314 | + | Accessory Gene | Resolvase |
| tnpA | Tn1721.1 | 9318-12284 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | mcp | Mcp | Tn1721.1 | 525 | 190-1767 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Comment: | methyl-accepting chemotaxis protein | ||||||||||
| Protein Sequence: | MRQNLPVTGR NLELPKDANI LSTTSPQSHI TYVNPDFIKI SGFTEEELLG QPHNIVRHPD MPPAAFEHMW STLKSGRSWM GLVKNRCKNG DHYWVSAYVT PIAKNGSIVE YQSVRTKPEP EQVLAAEKLY AQLRSGKAAR PKLAASFSVK ILLLIWGSII SSAMAAGMLT DTSISSLLLA TLMSGSLSSV SVLAILSPLG RLVERARNIS NNPLSQSLYT GRTDEFGQIE FALRMMQAET GAIVGRIGDA SNRLSEHTRG LLKDIESSNV LTVEQQAETD QIATAVNQMV ASIQEVASNA QHAADAAGRA DTETASGQRL VAHTSQSITA LEGEIRQATQ VIHELEGQSN EISKVLDVIR GIAEQTNLLA LNAAIEAARA GEQGRGFAVV ADEVRSLAAR TQQSTTDIQS MISALQERAQ SAVTVMEQSS RQAHTSVAHA EEAATALDGI GQRVNEITDM NAQIATAVEQ QGAVSEDINR SIINIRDAAD TNVQTGQNNL QSAKSVAQLT SALSELAKQF WEKRG |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniA | TniA | In_Tn1721.1 | 559 | 2073-3752 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | can be extended upstream by 12 amino acids| identical to tniA (Tn1721 and In2)| 25% amino acid sequence identity to TnsB from Tn7 | ||||||||||
| Protein Sequence: | MATDTPRIPE QGVATLPDEA WERARRRAEI ISPLAQSETV GHEAADMAAQ ALGLSRRQVY VLIRRARQGS GLVTDLVPGQ SGGGKGKGRL PEPVERVIHE LLQKRFLTKQ KRSLAAFHRE VTQVCKAQKL RVPARNTVAL RIASLDPRKV IRRREGQDAA RDLQGVGGEP PAVTAPLEQV QIDHTVIDLI VVDDRDRQPI GRPYLTLAID VFTRCVLGMV VTLEAPSAVS VGLCLVHVAC DKRPWLEGLN VEMDWQMSGK PLLLYLDNAA EFKSEALRRG CEQHGIRLDY RPLGQPHYGG IVERIIGTAM QMIHDELPGT TFSNPDQRGD YDSENKAALT LRELERWLTL AVGTYHGSVH NGLLQPPAAR WAEAVARVGV PAVVTRATSF LVDFLPILRR TLTRTGFVID HIHYYADALK PWIARRERWP SFLIRRDPRD ISRIWVLEPE GQHYLEIPYR TLSHPAVTLW EQRQALAKLR QQGREQVDES ALFRMIGQMR EIVTSAQKAT RKARRDADRR QHLKTSARPD KPVPPDTDIA DPQADNLPPA KPFDQIEEW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniB | TniB | In_Tn1721.1 | 302 | 3755-4663 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Sequence Family: | ATP binding protein? | ||||||||||
| Comment: | identical to tniB (Tn1721)| similar function to Tn7 tnsC and MuB | ||||||||||
| Protein Sequence: | MDEYPIIDLS HLLPAAQGLA RLPADERIQR LRADRWIGYP RAVEALNRLE ALYAWPNKQR MPNLLLVGPT NNGKSMIVEK FRRTHPASSD ADQEHIPVLV VQMPSEPSVI RFYVALLAAM GAPLRPRPRL PEMEQLALAL LRKVGVRMLV IDELHNVLAG NSVNRREFLN LLRFLGNELR IPLVGVGTRD AYLAIRSDDQ LENRFEPMML PVWEANDDCC SLLASFAASL PLRRPSPIAT LDMARYLLTR SEGTIGELAH LLMAAAIVAV ESGEEAINHR TLSMAVYTGP SERRRQFERE LM |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniQ | TniQ | In_Tn1721.1 | 405 | 4660-5877 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Target Site Selection | ||||||||||
| Comment: | identical to tniQ (Tn1721)|similar function to Tn7 tnsD? | ||||||||||
| Protein Sequence: | MKPAPRWPLH PAPKEGEALS SWLNRVALCY HMEEPDLLEH DLGHGQVDDL DTAPPLSLLA LLSQRSGIEL DRLRCMSFAG WVPWLLDSLD DQIPDALETY AFQLSVLLPR LRRKTRSITS WRAWLPSQPI NRACPLCLSD PENQAVLLAW KLPLMLSCPL HGCWLESYWG VPGRFLGWEN ADAEPRTASD AIAAMDQRTW QALTTGHVEL PRRRIHAGLW FRLLRTLLDE LNTPLSACGT CAGYPRQVWE GCGHPLRAGQ SLWRPYETLN PIVRLQMLEA AATAISLIEV RDISPPGEQA KLFWSEPQTG FTSGLPTKAP KPEPINHWQR AVQAIDEAII EARHNPETAR SLFALASYGR RDPASLERLR ATFVKEGIPP EFLSHYLPDA PFACLKQNDG LSDKF |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniR | TniR | In_Tn1721.1 | 207 | 5939-6562 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Comment: | resolution of cointegrates || Protein: ACE81792.1 || identical to tniR(Tn1721) | ||||||||||
| Protein Sequence: | MLIGYMRVSK ADGSQSTNLQ RDALIAAGVS LAHLYEDLAS GRRDDRPGLA ACLKALREGD TLIVWKLDRL GRDLRHLINT VHDLTARSVG LKVLTGHGAA VDTTTAAGKL VFGIFAALAE FERELISERT VAGLISARAR GRKGGRPFKM TAAKLRLAMA SMGQPETKVG DLCEELGITR QTLYRHVSPK GELRPDGVKL LSLGSAA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | qacE (ARO:3005009) | QacE | In_Tn1721.1 | 110 | 6811-7143 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic efflux (ARO:0010000) | ||||||||||
| Sequence Family: | major facilitator superfamily (MFS) antibiotic efflux pump (ARO:0010002) | ||||||||||
| Target: | disinfecting agents and antiseptics(ARO:3005386) | ||||||||||
| Comment: | SMR export pump||perfect match to reference sequence for ARO:3005009 (bitscore:207) | ||||||||||
| Protein Sequence: | MKGWLFLVIA IVGEVIATSA LKSSEGFTKL APSAVVIIGY GIAFYFLSLV MKSIPVGVAY ALWSGLGVVI ITAIAWLLHG QKLDAWGFVG MGLIVSGVVV LNLLSKASAH |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | intI1 | IntI1 | In_Tn1721.1 | 337 | 7388-8401 | + |
|---|
| Class: | Integron Integrase | ||||||||||
| Subclass: | Class 1 | ||||||||||
| Sequence Family: | Class 1 Integron Tyrosine Integrase | ||||||||||
| Transposase Chemistry: | Tyrosine | ||||||||||
| Protein Sequence: | MKTATAPLPP LRSVKVLDSC VSRIRYLHYS LRTEQAYVHW VRAFIRFHGV RHPATLGSSE VEAFLSWLAN ERKVSVSTHR QALAALLFFY GKVLCTDLPW LQEIGRPRPS RRLPVVLTPD EVVRILGFLE GEHRLFAQLL YGTGMRISEG LQLRVKDLDF DHGTIIVREG KGSKDRALML PESLAPSLRE QLSRARAWWL KDQAEGRSGV ALPDALERKY PRAGHSWPWF WVFAQHTHST DPRSGVVRRH HMYDQTFQRA FKRAVEQAGI TKPATPHTLR HSFATALLRS GYDIRTVQDL LGHSDVSTTM IYTHVLKVGG AGVRSPLDAL PPLTSER |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn1721.1 | 186 | 8754-9314 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MQGHRIGYVR VSSFDQNPER QLEQTQVSKV FTDKASGKDT QRPQLEALLS FVREGDTVVV HSMDRLARNL DDLRRLVQKL TQRGVRIEFL KEGLVFTGED SPMTNLMLSV MGAFAEFERA LIRERQREGI ALAKQRGAYR GRKKALSDEQ TVTLRQRAAA GEPKAQLARE FNISRETLYQ YLRTDD |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn1721.1 | 988 | 9318-12284 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MPRRLILSAT ERDTLLALPE SQDDLIRYYT FNDSDLSLIR QRRGDANRLG FAVQLCLLRY PGYALGTDSE LPEPVILWVA KQVQTDPASW TKYGERDVTR REHAQELRTY LQLAPFGLSD FRALVRELTK LAQQTDKGLL LAGQALESLR QKRRILPALS VIDRACSEAI ARANRRVYRA LVEPLTDSHR AKLDELLKLK AGSSITWLTW LRQAPLKPNS RHMLEHIERL KTFQLVDLPE GLGRHIHQNR LLKLAREGGQ MTPKDLGKFE PQRRYATLAA VVLESTATVI DELVDLHDRI LVKLFSGAKH KHQQQFQKQG KAINDKVRLY SKIGQALLEA KESGSDPYAA IEAVIPWDEF TESVSEAELL ARPEGFDHLH LVGENFATLR RYTPALLEVL ELRAAPAAQG VLAAVQTLRE MNADNLRKVP ADAPTAFIKP RWKPLVITPE GLDRRFYEIC ALSELKNALR SGDIWVKGSR QFRDFDDYLL PAEKFAALKR EQALPLAINP NSDQYLEERL QLLDEQLATV ARLAKDNDLP DAILTESGLK ITPLDAAVPD RAQALIDQTS QLLPRIKITE LLMDVDDWTG FSRHFTHLKD GAEAKDRTLL LSAILGDAIN LGLTKMAESS PGLTYAKLSW LQAWHIRDET YSAALAELVN HQYRHAFAAH WGDGTTSSSD GQRFRAGGRG ESTGHVNPKY GSEPGRLFYT HISDQYAPFS TRVVNVGVRD STYVLDGLLY HESDLRIEEH YTDTAGFTDH VFALMHLLGF RFAPRIRDLG ETKLYVPQGV QAYPTLRPLI GGTLNIKHVR AHWDDILRLA SSIKQGTVTA SLMLRKLGSY PRQNGLAVAL RELGRIERTL FILDWLQSVE LRRRVHAGLN KGEARNSLAR AVFFNRLGEI RDRSFEQQRY RASGLNLVTA AIVLWNTVYL ERATQGLVEA GKPVDGELLQ FLSPLGWEHI NLTGDYVWRQ SRRLEDGKFR PLRMPGKP |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| In_Tn1721.1-HQ730118.1 | In_Tn1721.1 | integron | 1932-8603 | + | 6672 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRt | In_Tn1721.1 | 1932-1964 | + | 33 | TGTCATTTTC AGAAGACGAC TGCACCAGTT GAT |
| repeat t1 | In_Tn1721.1 | 1940-1958 | + | 19 | TCAGAAGACG ACTGCACCA |
| repeat t2 | In_Tn1721.1 | 1980-1998 | - | 19 | TCAGGAGCTG GCTGCACAA |
| repeat t4 | In_Tn1721.1 | 2041-2059 | + | 19 | TCAATACTCG TGTGCACCA |
| repeat i4 | In_Tn1721.1 | 8484-8502 | - | 19 | TCAGCGGACG CAGGGAGGA |
| repeat i3 | In_Tn1721.1 | 8512-8530 | - | 19 | TCAGGCAACG ACGGGCTGC |
| repeat i2 | In_Tn1721.1 | 8554-8572 | - | 19 | TCAGAAGCCG ACTGCACTA |
| IRi | In_Tn1721.1 | 8571-8603 | - | 33 | TGTCGTTTTC AGAAGACGGC TGCACTGAAC GTC |
| repeat i1 | In_Tn1721.1 | 8577-8595 | - | 19 | TCAGAAGACG GCTGCACTG |
1. Betteridge T, Partridge SR, Iredell JR, Stokes HW.
Genetic context and structural diversity of class 1 integrons from human commensal bacteria in a hospital intensive care unit.. Antimicrob Agents Chemother. 2011 Aug;55(8):3939-43. doi: 10.1128/AAC.01831-10. Epub 2011 May 31. PubMed ID:
21628540
.