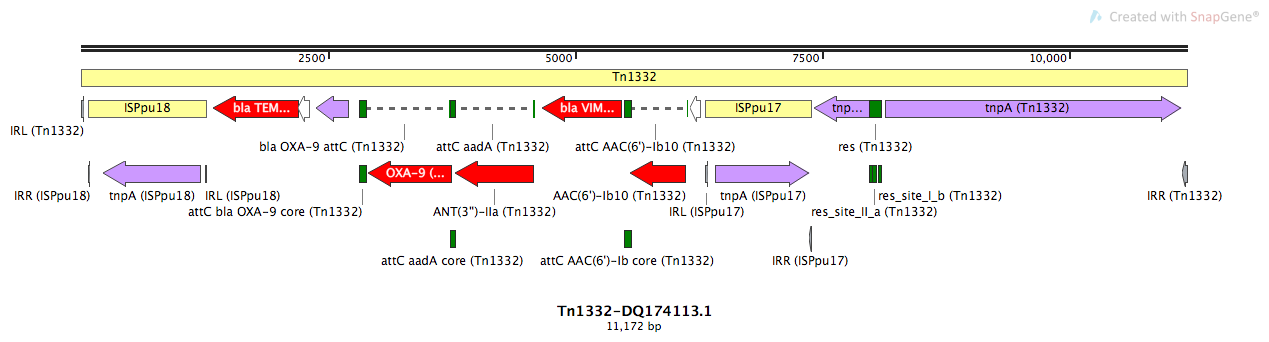
Mobile element type:
Transposon
Name:
Tn1332
Synonyms:
Accession:
Tn1332-DQ174113.1
Family:
Tn3
Group:
Tn3
First isolate:
Yes
Partial:
ND
Evidence of transposition:
ND
Host Organism:
Pseudomonas putida 9335
Date of Isolation:
January 2004
Country:
France
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-38 | + | 38 | GGGGTCTGAC GCTCAGTGGA ACGAAAACTC ACGTTAAG |
| IRR | 11135-11172 | - | 38 | GGGGTCTGAC GCTCAGTGGA ACGAAAACTC ACGTTAAG |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| attC bla OXA-9 core | Tn1332 | 2818-2886 | + | 69 | CGTTCAAGGT GAGCGGACTC GCGCGGCTTT ATGCGCGAGG TCCGCTCGAG CGCAGGGTTA TGCATCTGC |
| attC aadA | Tn1332 | 3733-3787 , 4581-4586 | + | 61 | GGTATCCTTC ATTGCTCCGC TGTGCGCTTA ATAGGTGCAT AACTTTGTTT TAGACGTTTA A |
| attC aadA core | Tn1332 | 3733-3787 | + | 55 | GGTATCCTTC ATTGCTCCGC TGTGCGCTTA ATAGGTGCAT AACTTTGTTT TAGAC |
| attC AAC(6')-Ib10 | Tn1332 | 5496-5561 , 6129-6135 | + | 73 | CGTTTGACAT GAGGGGCGGC CAAGGGCGCC AGCCCTTGGA CGTCCCCCTC GATGGAAGGG TTAGGCGCCT AAC |
| attC AAC(6')-Ib core | Tn1332 | 5496-5561 | + | 66 | CGTTTGACAT GAGGGGCGGC CAAGGGCGCC AGCCCTTGGA CGTCCCCCTC GATGGAAGGG TTAGGC |
| res | Tn1332 | 7973-8093 | + | 121 | AAATGTACCT TAAATCGAAT ATCAGACACG ATGTGTCTAT TATGCCAAAA TGACGATTTA ATGGACACTC AAACGAAGCC GTTTTACTAT GTCTGATAAT TTATAACATT TCGTACGGTT G |
| res_site_III_a | Tn1332 | 7976-7998 | + | 23 | TGTACCTTAA ATCGAATATC AGA |
| res_site_II_a | Tn1332 | 8004-8039 | + | 36 | TGTGTCTATT ATGCCAAAAT GACGATTTAA TGGACA |
| res_site_I_b | Tn1332 | 8062-8090 | + | 29 | TGTCTGATAA TTTATAACAT TTCGTACGG |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| tnpA | ISPpu18 | 226-1206 | - | Transposase | |
| bla TEM-1 (ARO:3000873) | Tn1332 | 1344-2204 | - | Passenger Gene | Antibiotic Resistance |
| tnpR_p | Tn1332 | 2387-2705 | - | Accessory Gene | Resolvase |
| OXA-9 (ARO:3001404) | Tn1332 | 2905-3744 | - | Passenger Gene | Antibiotic Resistance |
| ANT(3'')-IIa (ARO:3004089) | Tn1332 | 3789-4577 | - | Passenger Gene | Antibiotic Resistance |
| bla VIM-17 (ARO:3002287) | Tn1332 | 4665-5465 | - | Passenger Gene | Antibiotic Resistance |
| AAC(6')-Ib10 (ARO:3002581) | Tn1332 | 5556-6110 | - | Passenger Gene | Antibiotic Resistance |
| tnpA | ISPpu17 | 6415-7377 | + | Transposase | |
| tnpR | Tn1332 | 7413-7970 | - | Accessory Gene | Resolvase |
| tnpA | Tn1332 | 8134-11139 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | ISPpu18 | 326 | 226-1206 | - |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MKQMTFADAE YAGKRKQTRK ELFLIEMDQV VPWKGLIALI EPHYPKGEGG RPAYPLMAML RIHLMQNWFG YSDPAMEEAL YETTILRQFA GLSLERIPDE TTILKFRRLL EKHELAAGIL GVINGYLGDR GLSLRQGTIV DATLIHAPSS TKNQDGKRDP EMHQSKKGNQ YYFGMKAHIG VDDESGLVHS VVGTAANVAD ITQVDKLLHG DENVVCADAG YTGVEKRPEH AGREVIWQVA ARRSTYKKLD KRSALYKAKR KIEKAKAQVR AKVEHPFRVI KRQFGYVKVR FRGLAKNTAQ LVTLFALSNL WMARRHLLTT AGKVHL |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | bla TEM-1 (ARO:3000873) | Bla TEM-1 | Tn1332 | 286 | 1344-2204 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | TEM beta-lactamase (ARO:3000014) | ||||||||||
| Target: | penem (ARO:3003706)||cephalosporin (ARO:0000032)||monobactam (ARO:0000004)||penam (ARO:3000008) | ||||||||||
| Comment: | perfect match to reference sequence for ARO:3000873||Synonyms: TEM-98, RTEM-1 | ||||||||||
| Protein Sequence: | MSIQHFRVAL IPFFAAFCLP VFAHPETLVK VKDAEDQLGA RVGYIELDLN SGKILESFRP EERFPMMSTF KVLLCGAVLS RVDAGQEQLG RRIHYSQNDL VEYSPVTEKH LTDGMTVREL CSAAITMSDN TAANLLLTTI GGPKELTAFL HNMGDHVTRL DRWEPELNEA IPNDERDTTM PAAMATTLRK LLTGELLTLA SRQQLIDWME ADKVAGPLLR SALPAGWFIA DKSGAGERGS RGIIAALGPD GKPSRIVVIY TTGSQATMDE RNRQIAEIGA SLIKHW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR_p | TnpR_p | Tn1332 | 106 | 2387-2705 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Comment: | C-terminal fragment | ||||||||||
| Protein Sequence: | KRV*CSGCSG SVY*RRDQYR R*YGANGGHH PVGCGTG*TP EDPRAHE*GP TGSKAERNQI WPQAYRGQER RADASSEGHW CNGNCSSAQY CPLHGL*NS* RRKGLV |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | OXA-9 (ARO:3001404) | OXA-9 | Tn1332 | 279 | 2905-3744 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | OXA beta-lactamase (ARO:3000017) | ||||||||||
| Target: | cephalosporin (ARO:0000032)||penam (ARO:3000008) | ||||||||||
| Comment: | perfect match to reference sequence for ARO:3001404 | ||||||||||
| Protein Sequence: | MKDTLMKKIL LLHMLVFVSA TLPISSVASD EVETLKCTII ADAITGNTLY ETGECARRVS PCSSFKLPLA IMGFDSGILQ SPKSPTWELK PEYNPSPRDR TYKQVYPALW QSDSVVWFSQ QLTSRLGVDR FTEYVKKFEY GNQDVSGDSG KHNGLTQSWL MSSLTISPKE QIQFLLRFVA HKLPVSEAAY DMAYATIPQY QAAEGWAVHG KSGSGWLRDN NGKINESRPQ GWFVGWAEKN GRQVVFARLE IGKEKSDIPG GSKAREDILV ELPVLMGNK |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | ANT(3'')-IIa (ARO:3004089) | ANT(3'')-IIa | Tn1332 | 262 | 3789-4577 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | ANT(3'')(ARO:3004275) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | strict match to reference sequence for ARO:3004089 (bitscore: 569)||Synonym: ANT(9)(3'') | ||||||||||
| Protein Sequence: | MREAVIAEVS TQLSEVVGVI ERHLEPTLLA VHLYGSAVDG GLKPHSDIDL LVTVTVRLDE TTRRALINDL LETSASPGES EILRAVEVTI VVHDDIIPWR YPAKRELQFG EWQRNDILAG IFEPATIDID LAILLTKARE HSVALVGPAA EELFDPVPEQ DLFEALNETL TLWNSPPDWA GDERNVVLTL SRIWYSAVTG KIAPKDVAAD WAMERLPAQY QPVILEARQA YLGQEDRLAS RADQLEEFVH YVKGEITKVV GK |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | bla VIM-17 (ARO:3002287) | Bla VIM-17 | Tn1332 | 266 | 4665-5465 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | VIM beta-lactamase (ARO:3000021) | ||||||||||
| Target: | cephamycin (ARO:0000044)||cephalosporin (ARO:0000032)||penam (ARO:3000008)||carbapenem (ARO:0000020)||penem (ARO:3003706) | ||||||||||
| Comment: | strict match to reference sequence for ARO:3002287 (bitscore: 538) | ||||||||||
| Protein Sequence: | MFKLLSKLLV YLTASIMAIA SPLAFSVDSS GEYPTVSEIP VGEVRLYQIA DGVWSHIATQ SFDGAVYPSN GLIVRDGDEL LLIDTAWGAK NTAALLAEIE KQIGLPVTRA VSTHFHDDRV GGVDVLRAAG VATYASPSTR RLAEVEGNEI PTHSLEGLSS SGDAVRFGPV ELFYPGAAHS TDNLVVYVPS ASVLYGGCAI YELSRTSAGN VADADLAEWP TSIERIQQHY PEAQFVIPGH GLPGGLDLLK HTTNVVKAHT NRSVVE |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | AAC(6')-Ib10 (ARO:3002581) | AAC(6')-Ib10 | Tn1332 | 184 | 5556-6110 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | AAC(6') (ARO:3000345) | ||||||||||
| Transposase Chemistry: | aminoglycoside acetyltransferase | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | strict match to reference sequence for ARO:3002581 (bitscore: 377) | ||||||||||
| Protein Sequence: | MTNSNDSVTL RLMTEHDLAM LYEWLNRSHI VEWWGGEEAR PTLADVQEQY LPSVLAQESV TPYIAMLNGE PIGYAQSYVA LGSGDGWWEE ETDPGVRGID QLLANASQLG KGLGTKLVRA LVELLFNDPE VTKIQTDPSP SNLRAIRCYE KAGFERQGTV TTPDGPAVYM VQTRQAFERT RSVA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | ISPpu17 | 320 | 6415-7377 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MTTHYRQLTQ GQRYQIEAGL SAAESQASIA KRVGVHPSTI SREVRRNSTQ NIYKAVSAAH ESDARRAGAR KFCKPATWLS HHLPVWLKHG MSPEQIAQRL KQEQPSRAVS HEWIYRFIAA EQRAGGELYT YLRHRRKRYR KRYGSHDRRG QLRNRVSISE RPAEVESRER LGDWEGDTVH GLGGNLVTLV DRKSGYLSAY PVKRRTRRQV TRAINLMLQG HAAHTLTLDN GREFAGHERI ALRSQCQVFF ADPYSSWQRG TNENTNGLLR QYFPKGSDFS KLTVEAVNRT VARINLRPRK RLGWKTPYEV HTGVSVALMC |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpR | TnpR | Tn1332 | 185 | 7413-7970 | - |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Resolvase | ||||||||||
| Function: | resolvase||serine site-specific recombinase | ||||||||||
| Sequence Family: | Serine Site-Specific Recombinase | ||||||||||
| Transposase Chemistry: | Serine | ||||||||||
| Protein Sequence: | MRIFGYARVS TSQQSLDIQI RALKDAGVKA NRIFTDKASG SSTDREGLDL LRMKVEEGDV ILVKKLDRLG RDTADMIQLI KEFDAQGVAV RFIDDGISTD GDMGQMVVTI LSAVAQAERR RILERTNEGR QEAKLKGIKF GRRRTVDRNV VLTLHQKGTG ATEIAHQLSI ARSTVYKILE DERAS |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnpA | TnpA | Tn1332 | 1001 | 8134-11139 | + |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MPVDFLTTEQ TESYGRFTGE PDELQLARYF HLDEADKEFI GKSRGDHNRL GIALQIGCVR FLGTFLTDMN HIPSGVRHFT ARQLGIRDIT VLAEYGQREN TRREHAALIR QHYQYREFAW PWTFRLTRLL YTRSWISNER PGLLFDLATG WLMQHRIILP GATTLTRLIS EVREKATLRL WNKLALIPSA EQRSQLEMLL GPTDCSRLSL LESLKKGPVT ISGPAFNEAI ERWKTLNDFG LHAENLSTLP AVRLKNLARY AGMTSVFNIA RMSPQKRMAV LVAFVLAWET LALDDALDVL DAMLAVIIRD ARKIGQKKRL RSLKDLDKSA LALASACSYL LKEETPDESI RAEVFSYIPR QKLAEIITLV REIARPSDDN FHEEMVEQYG RVRRFLPHLL NTVKFSSAPA GVTTLNACDY LSREFSSRRQ FFDDAPTEII SRSWKRLVIN KEKHITRRGY TLCFLSKLQD SLRRRDVYVT GSNRWGDPRA RLLQGADWQA NRIKVYRSLG HPTDPQEAIK SLGHQLDSRY RQVAARLGEN EAVELDVSGP KPRLTISPLA SLDEPDSLKR LSKMISDLLP PVDLTELLLE INAHTGFADE FFHASEASAR VDDLPVSISA VLMAEACNIG LEPLIRSNVP ALTRHRLNWT KANYLRAETI TSANARLVDF QATLPLAQIW GGGEVASADG MRFVTPVRTI NAGPNRKYFG NNRGITWYNF VSDQYSGFHG IVIPGTLRDS IFVLEGLLEQ ETGLNPTEIM TDTAGASDLV FGLFWLLGYQ FSPRLADAGA SVFWRMDHDA DYGVLNDIAR GQSDPRKIVL QWDEMIRTAG SLKLGKVQAS VLVRSLLKSE RPSGLTQAII EVGRINKTLY LLNYIDDEDY RRRILTQLNR GESRHAVARA ICHGQKGEIR KRYTDGQEDQ LGALGLVTNA VVLWNTIYMQ AALDHLRAQG ETLNDEDIAR LSPLCHGHIN MLGHYSFTLA ELVTKGHLRP LKEASEAENV A |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| ISPpu18-DQ174113.1 | ISPpu18 | insertion sequence | 80-1271 | + | 1192 |
| ISPpu17-DQ174113.1 | ISPpu17 | insertion sequence | 6319-7384 | + | 1066 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRR | ISPpu18 | 80-91 | + | 12 | GGAAGGTCTG AA |
| IRL | ISPpu18 | 1260-1271 | - | 12 | GGAAGGTCTG AA |
| IRL | ISPpu17 | 6319-6340 | + | 22 | CCTGAATTCA TAGAATAAGC GC |
| IRR | ISPpu17 | 7363-7384 | - | 22 | CCTGAATTCA ACACATAAGT GC |
1. Poirel L, Cabanne L, Collet L, Nordmann P.
Class II transposon-borne structure harboring metallo-beta-lactamase gene blaVIM-2 in Pseudomonas putida.. Antimicrob Agents Chemother. 2006 Aug;50(8):2889-91. doi: 10.1128/AAC.00398-06. PubMed ID:
16870796
.