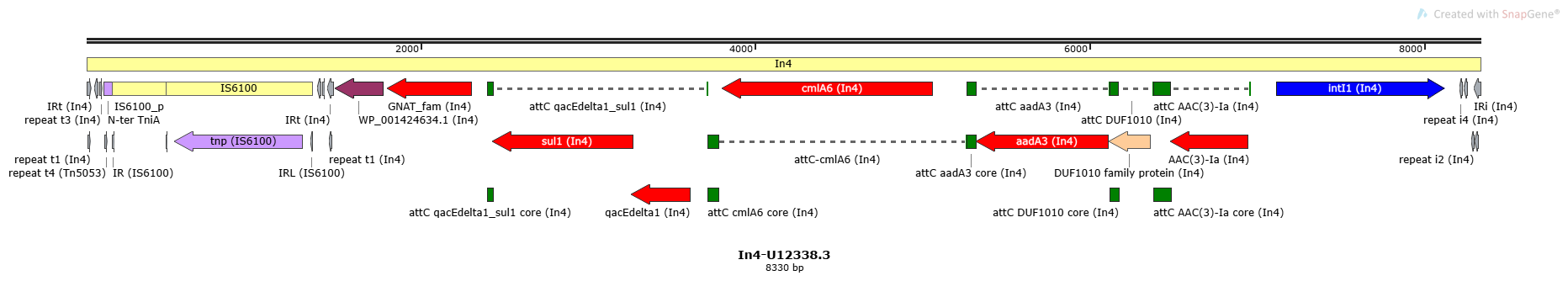
Mobile element type:
Integron
Name:
In4
Synonyms:
Accession:
In4-U12338.3
Family:
Tn402
Group:
Class 1
First isolate:
Yes
Partial:
ND
Evidence of transposition:
ND
Host Organism:
Pseudomonas aeuroginosa
Date of Isolation:
Country:
Spain
Molecular Source:
plasmid R1033
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRt | 1-33 | + | 33 | TGTCATTTTC AGAAGACGAC TGCACCAGTT GAT |
| repeat t1 | 9-27 | + | 19 | TCAGAAGACG ACTGCACCA |
| repeat t2 | 49-67 | - | 19 | TCAGGAGCTG GCTGCACAA |
| repeat t3 | 78-97 | + | 20 | TCAGAAGTGA TCTGCACCAA |
| repeat t3 | 1380-1399 | - | 20 | TCAGAAGTGA TCTGCACCAA |
| repeat t2 | 1410-1428 | + | 19 | TCAGGAGCTG GCTGCACAA |
| IRt | 1444-1476 | - | 33 | TGTCATTTTC AGAAGACGAC TGCACCAGTT GAT |
| repeat t1 | 1450-1468 | - | 19 | TCAGAAGACG ACTGCACCA |
| repeat i4 | 8211-8229 | - | 19 | TCAGCGGACG CAGGGAGGA |
| repeat i3 | 8239-8257 | - | 19 | TCAGGCAACG ACGGGCTGC |
| repeat i2 | 8281-8299 | - | 19 | TCAGAAGCCG ACTGCACTA |
| IRi | 8298-8330 | - | 33 | TGTCGTTTTC AGAAGACGGC TGCACTGAAC GTC |
| repeat i1 | 8304-8322 | - | 19 | TCAGAAGACG GCTGCACTG |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| attC qacEdelta1_sul1 | In4 | 2399-2432 , 3710-3716 | + | 41 | CCGCTAGCGG GCGGCCGGAA GGTGAATGCT AGGCATCTAA C |
| attC qacEdelta1_sul1 core | In4 | 2399-2432 | + | 34 | CCGCTAGCGG GCGGCCGGAA GGTGAATGCT AGGC |
| attC-cmlA6 | In4 | 3716-3779 , 5259-5264 | + | 70 | CGCCTGAGCT CAGCCGACCG AAACCGCGTA GCGGTTTTGG GTCGGCTGCA GCGATTTGTT GGGCGCCCAA |
| attC-cmlA6 5'-end | In4 | 3716-3779 | + | 64 | CGCCTGAGCT CAGCCGACCG AAACCGCGTA GCGGTTTTGG GTCGGCTGCA GCGATTTGTT GGGC |
| attC cmlA6 core | In4 | 3716-3779 | + | 64 | CGCCTGAGCT CAGCCGACCG AAACCGCGTA GCGGTTTTGG GTCGGCTGCA GCGATTTGTT GGGC |
| attC-cmlA6 3'-end | In4 | 5259-5264 | + | 6 | GCCCAA |
| attC aadA3 | In4 | 5265-5318 , 6115-6120 | + | 60 | CGCCGGAGTT AAGCCGCCGC GCGTAGCGCG GTCGGCTTGA ACGAATTGTT AGACGTCTAA |
| attC-aadA3 5'-end | In4 | 5265-5318 | + | 54 | CGCCGGAGTT AAGCCGCCGC GCGTAGCGCG GTCGGCTTGA ACGAATTGTT AGAC |
| attC aadA3 core | In4 | 5265-5318 | + | 54 | CGCCGGAGTT AAGCCGCCGC GCGTAGCGCG GTCGGCTTGA ACGAATTGTT AGAC |
| attC-aadA3 3'-end | In4 | 6115-6120 | + | 6 | GTCTAA |
| attC DUF1010 | In4 | 6121-6174 , 6377-6382 | + | 60 | CGGCCAAGGT AAGCGGCCCG CAGAATGCGG GTCCGCTTGA CCGCAGAGTT AGACGACTAA |
| attC-DUF1010 5'-end | In4 | 6121-6174 | + | 54 | CGGCCAAGGT AAGCGGCCCG CAGAATGCGG GTCCGCTTGA CCGCAGAGTT AGAC |
| attC DUF1010 core | In4 | 6121-6174 | + | 54 | CGGCCAAGGT AAGCGGCCCG CAGAATGCGG GTCCGCTTGA CCGCAGAGTT AGAC |
| attC-DUF1010 3'-end | In4 | 6377-6382 | + | 6 | GACTAA |
| attC AAC(3)-Ia | In4 | 6383-6485 , 6954-6959 | + | 109 | CGCCTGAGTT AAGCCGGAGC GCTTTGCGGC CGCGGCGTTG TGACAATTTA CCGAACAACT CCGCGGCCGG GAAGCCGATC TCGGCTTGAA CGAATTGTTA GGTACCTAA |
| attC-AAC(3)-Ia 5'-end | In4 | 6383-6485 | + | 103 | CGCCTGAGTT AAGCCGGAGC GCTTTGCGGC CGCGGCGTTG TGACAATTTA CCGAACAACT CCGCGGCCGG GAAGCCGATC TCGGCTTGAA CGAATTGTTA GGT |
| attC AAC(3)-Ia core | In4 | 6383-6485 | + | 103 | CGCCTGAGTT AAGCCGGAGC GCTTTGCGGC CGCGGCGTTG TGACAATTTA CCGAACAACT CCGCGGCCGG GAAGCCGATC TCGGCTTGAA CGAATTGTTA GGT |
| attC-AAC(3)-Ia 3'-end | In4 | 6954-6959 | + | 6 | ACCTAA |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| tnp | IS6100 | 528-1292 | - | Transposase | |
| WP_001424634.1 | In4 | 1488-1775 | - | Passenger Gene | Hypothetical |
| GNAT_fam | In4 | 1799-2299 | - | Passenger Gene | Antibiotic Resistance |
| sul1 (ARO:3000410) | In4 | 2427-3266 | - | Passenger Gene | Antibiotic Resistance |
| qacEdelta1 (ARO:3005010) | In4 | 3260-3607 | - | Passenger Gene | Antibiotic Resistance |
| cmlA6 (ARO:3002696) | In4 | 3799-5058 | - | Passenger Gene | Antibiotic Resistance |
| aadA3 (ARO:3002603) | In4 | 5320-6111 | - | Passenger Gene | Antibiotic Resistance |
| DUF1010 family protein | In4 | 6117-6362 | - | Passenger Gene | Other |
| AAC(3)-Ia (ARO:3002528) | In4 | 6480-6944 | - | Passenger Gene | Antibiotic Resistance |
| intI1 | In4 | 7115-8128 | + | Integron Integrase | Class 1 |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tnp | Tnp | IS6100 | 254 | 528-1292 | - |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MTDFKWRHFQ GDVILWAVRW YCRYPISYRD LEEMLAERGI SVDHTTIYRW VQCYAPEMEK RLRWFWRRGF DPSWRLDETY VKVRGKWTYL YRAVDKRGDT IDFYLSPTRS AKAAKRFLGK ALRGLKHWEK PATLNTDKAP SYGAAITELK REGKLDRETA HRQVKYLNNV IEADHGKLKI LIKPVRGFKS IPTAYATIKG FEVMRALRKG QARPWCLQPG IRGEVRLVER AFGIGPSALT EAMGMLNHHF AAAA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | WP_001424634.1 | WP_001424634.1 | In4 | 95 | 1488-1775 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Hypothetical | ||||||||||
| Protein Sequence: | MSGWDGPVAS RSLAVRPRCL GCARGCRYPP LPSSTLDNAA AVIAADARAG LFGPVRLARP SACLGEISGG RINGGASPPF VRVKAEDSSL KNIRL |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | GNAT_fam | GNAT_fam | In4 | 166 | 1799-2299 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Sequence Family: | GNAT | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Protein Sequence: | MDSEEPPNVR VACSGDIDEV VRLMHDAAAW MSAKGTPAWD VARIDRTFAE TFVLRSELLV ASCSDGIVGC CTLSAEDPEF WPDALKGEAA YLHKLAVRRT HAGRGVSSAL IEACRHAART QGCAKLRLDC HPNLRGLYER LGFTHVDTFN PGWDPTFIAE RLELEI |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | sul1 (ARO:3000410) | Sul1 | In4 | 279 | 2427-3266 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic target replacement (ARO:0001002) | ||||||||||
| Sequence Family: | sulfonamide resistant sul (ARO:3004238) | ||||||||||
| Transposase Chemistry: | dihydropteroate synthase | ||||||||||
| Target: | sulfonamide antibiotic (ARO:3000282)||sulfone antibiotic (ARO:3003401) | ||||||||||
| Comment: | perfect match to reference sequence for ARO:3000410 | ||||||||||
| Protein Sequence: | MVTVFGILNL TEDSFFDESR RLDPAGAVTA AIEMLRVGSD VVDVGPAASH PDARPVSPAD EIRRIAPLLD ALSDQMHRVS IDSFQPETQR YALKRGVGYL NDIQGFPDPA LYPDIAEADC RLVVMHSAQR DGIATRTGHL RPEDALDEIV RFFEARVSAL RRSGVAADRL ILDPGMGFFL SPAPETSLHV LSNLQKLKSA LGLPLLVSVS RKSFLGATVG LPVKDLGPAS LAAELHAIGN GADYVRTHAP GDLRSAITFS ETLAKFRSRD ARDRGLDHA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | qacEdelta1 (ARO:3005010) | QacEdelta1 | In4 | 115 | 3260-3607 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic efflux (ARO:0010000) | ||||||||||
| Sequence Family: | major facilitator superfamily (MFS) antibiotic efflux pump (ARO:0010002) | ||||||||||
| Target: | disinfecting agents and antiseptics(ARO:3005386) | ||||||||||
| Comment: | subunit of the qac multidrug efflux pump||perfect match to reference sequence for ARO:3005010 (bitscore:219) | ||||||||||
| Protein Sequence: | MKGWLFLVIA IVGEVIATSA LKSSEGFTKL APSAVVIIGY GIAFYFLSLV LKSIPVGVAY AVWSGLGVVI ITAIAWLLHG QKLDAWGFVG MGLIIAAFLL ARSPSWKSLR RPTPW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | cmlA6 (ARO:3002696) | CmlA6 | In4 | 419 | 3799-5058 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic efflux (ARO:0010000) | ||||||||||
| Sequence Family: | major facilitator superfamily (MFS) antibiotic efflux pump (ARO:0010002) | ||||||||||
| Target: | phenicol antibiotic (ARO:3000387) | ||||||||||
| Comment: | strict match to reference sequence for ARO:3002696 (bitscore: 819) | ||||||||||
| Protein Sequence: | MSSKNFSWRY SLAATVLLLS PFDLLASLGM DMYLPAVPFM PNALGTTAST IQLTLTTYLV MIGAGQLLFG PLSDRLGRRP VLLGGGLAYV VASMGLALTS SAEVFLGLRI LQACGASACL VSTFATVRDI YAGREESNVI YGILGSMLAM VPAVGPLLGA LVDMWLGWRA IFAFLGLGMI AASAAAWRFW PETRVQRVAG LQWSQLLLPV KCLNFWLYTL CYAAGMGSFF VFFSIAPGLM MGRQGVSQLG FSLLFATVAI AMVFTARFMG RVIPKWGSPS VLRMGMGCLI AGAVLLAITE IWALQSVLGF IAPMWLVGIG VATAVSVAPN GALRGFDHVA GTVTAVYFCL GGVLLGSIGT LIISLLPRNT AWPVVVYCLT LATVVLGLSC VSRVKGSRGQ GEHDVVALQS AESTSNPNR |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | aadA3 (ARO:3002603) | AadA3 | In4 | 263 | 5320-6111 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | ANT(3'')(ARO:3004275) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | strict match to reference sequence for ARO:3002603 (bitscore: 522) | ||||||||||
| Protein Sequence: | MRVAVTIEIS NQLSEVLSVI ERHLESTLLA VHLYGSAVDG GLKPYSDIDL LVTVAVKLDE TTRRALLNDL MEASAFPGES ETLRAIEVTL VVHDDIIPWR YPAKRELQFG EWQRNDILAG IFEPAMIDID LAILLTKARE HSVALVGPAA EEFFDPVPEQ DLFEALRETL KLWNSQPDWA GDERNVVLTL SRIWYSAITG KIAPKDVAAD WAIKRLPAQY QPVLLEAKQA YLGQKEDHLA SRADHLEEFI RFVKGEIIKS VGK |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | DUF1010 family protein | DUF1010 family protein | In4 | 81 | 6117-6362 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Other | ||||||||||
| Sequence Family: | DUF1010 (Pfam:PF06231) | ||||||||||
| Comment: | BLAST: WP_032492246.1 | ||||||||||
| Protein Sequence: | MRAATYFSAP PCSKGFLAFW ASSACAASAT SYPSCSAAPL SWSSGFSWAA PFFKAGRSLL AFGSNSAVKR TRILRAAYLG R |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | AAC(3)-Ia (ARO:3002528) | AAC(3)-Ia | In4 | 154 | 6480-6944 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic inactivation (ARO:0001004) | ||||||||||
| Sequence Family: | AAC(3) (ARO:3000322) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | perfect match to reference sequence for ARO:3002528||Synonyms: AAC(3)-Ia C, aacC1, aacC-A1 | ||||||||||
| Protein Sequence: | MGIIRTCRLG PDQVKSMRAA LDLFGREFGD VATYSQHQPD SDYLGNLLRS KTFIALAAFD QEAVVGALAA YVLPRFEQPR SEIYIYDLAV SGEHRRQGIA TALINLLKHE ANALGAYVIY VQADYGDDPA VALYTKLGIR EEVMHFDIDP STAT |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | intI1 | IntI1 | In4 | 337 | 7115-8128 | + |
|---|
| Class: | Integron Integrase | ||||||||||
| Subclass: | Class 1 | ||||||||||
| Sequence Family: | Class 1 Integron Tyrosine Integrase | ||||||||||
| Transposase Chemistry: | Tyrosine | ||||||||||
| Protein Sequence: | MKTATAPLPP LRSVKVLDQL RERIRYLHYS LRTEQAYVNW VRAFIRFHGV RHPATLGSSE VEAFLSWLAN ERKVSVSTHR QALAALLFFY GKVLCTDLPW LQEIGRPRPS RRLPVVLTPD EVVRILGFLE GEHRLFAQLL YGTGMRISEG LQLRVKDLDF DHGTIIVREG KGSKDRALML PESLAPTLRE QLSRARAWWL KDQAEGRSGV ALPDALERKY PRAGHSWPWF WVFAQHTHST DPRSGVVRRH HMYDQTFQRA FKRAVEQAGI TKPATPHTLR HSFATALLRS GYDIRTVQDL LGHSDVSTTM IYTHVLKVGG AGVRSPLDAL PPLTSER |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| IS6100-U12338.3 | IS6100 | insertion sequence | 474-1353 | + | 880 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRR | IS6100 | 474-487 | + | 14 | GGCTCTGTTG CAAA |
| IRL | IS6100 | 1340-1353 | - | 14 | GGCTCTGTTG CAAA |
1. Wohlleben W, Arnold W, Bissonnette L, Pelletier A, Tanguay A, Roy PH, Gamboa GC, Barry GF, Aubert E, Davies J, et al..
On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacC1) for gentamicin acetyltransferase-3-I(AAC(3)-I), another member of the Tn21-based expression cassette.. Mol Gen Genet. 1989 Jun;217(2-3):202-8. doi: 10.1007/BF02464882. PubMed ID:
2549372
.
2. Bissonnette L, Champetier S, Buisson JP, Roy PH. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins.. J Bacteriol. 1991 Jul;173(14):4493-502. doi: 10.1128/jb.173.14.4493-4502.1991. PubMed ID: 1648560 .
3. Stokes HW, Hall RM. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation.. Plasmid. 1991 Jul;26(1):10-9. doi: 10.1016/0147-619x(91)90032-r. PubMed ID: 1658833 .
4. Collis CM, Hall RM. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase.. J Bacteriol. 1992 Mar;174(5):1574-85. doi: 10.1128/jb.174.5.1574-1585.1992. PubMed ID: 1311297 .
5. Hall RM, Brown HJ, Brookes DE, Stokes HW. Integrons found in different locations have identical 5' ends but variable 3' ends.. J Bacteriol. 1994 Oct;176(20):6286-94. doi: 10.1128/jb.176.20.6286-6294.1994. PubMed ID: 7929000 .
6. Partridge SR, Brown HJ, Stokes HW, Hall RM. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins.. Antimicrob Agents Chemother. 2001 Apr;45(4):1263-70. doi: 10.1128/AAC.45.4.1263-1270.2001. PubMed ID: 11257044 .
7. Smith DI, Lus RG, Rubio Calvo MC, Datta N, Jacob AE, Hedges RW. Third type of plasmid conferring gentamicin resistance in Pseudomonas aeruginosa.. Antimicrob Agents Chemother. 1975 Sep;8(3):227-30. doi: 10.1128/AAC.8.3.227. PubMed ID: 810080 .
2. Bissonnette L, Champetier S, Buisson JP, Roy PH. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins.. J Bacteriol. 1991 Jul;173(14):4493-502. doi: 10.1128/jb.173.14.4493-4502.1991. PubMed ID: 1648560 .
3. Stokes HW, Hall RM. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation.. Plasmid. 1991 Jul;26(1):10-9. doi: 10.1016/0147-619x(91)90032-r. PubMed ID: 1658833 .
4. Collis CM, Hall RM. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase.. J Bacteriol. 1992 Mar;174(5):1574-85. doi: 10.1128/jb.174.5.1574-1585.1992. PubMed ID: 1311297 .
5. Hall RM, Brown HJ, Brookes DE, Stokes HW. Integrons found in different locations have identical 5' ends but variable 3' ends.. J Bacteriol. 1994 Oct;176(20):6286-94. doi: 10.1128/jb.176.20.6286-6294.1994. PubMed ID: 7929000 .
6. Partridge SR, Brown HJ, Stokes HW, Hall RM. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins.. Antimicrob Agents Chemother. 2001 Apr;45(4):1263-70. doi: 10.1128/AAC.45.4.1263-1270.2001. PubMed ID: 11257044 .
7. Smith DI, Lus RG, Rubio Calvo MC, Datta N, Jacob AE, Hedges RW. Third type of plasmid conferring gentamicin resistance in Pseudomonas aeruginosa.. Antimicrob Agents Chemother. 1975 Sep;8(3):227-30. doi: 10.1128/AAC.8.3.227. PubMed ID: 810080 .