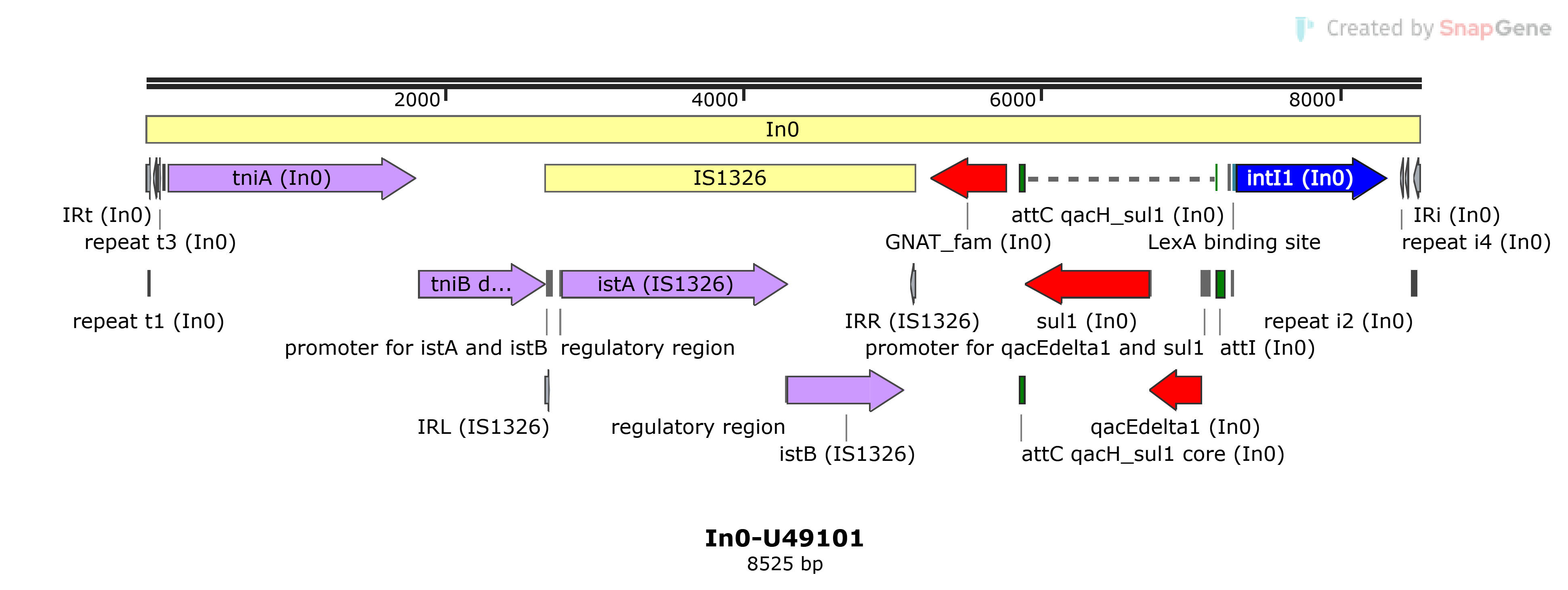
Mobile element type:
Integron
Name:
In0
Synonyms:
Accession:
In0-U49101
Family:
Tn402
Group:
Class 1
First isolate:
Yes
Partial:
ND
Evidence of transposition:
Yes
Host Organism:
Pseudomonas aeuroginosa PAT
Date of Isolation:
1992
Country:
Molecular Source:
plasmid pVS1
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRt | 1-33 | + | 33 | TGTCATTTTC AGAAGACGAC TGCACCAGTT GAT |
| repeat t1 | 9-27 | + | 19 | TCAGAAGACG ACTGCACCA |
| repeat t2 | 49-67 | - | 19 | TCAGGAGCTG GCTGCACAA |
| repeat t3 | 78-97 | + | 20 | TCAGAAGTGA TCTGCACCAA |
| repeat t4 | 110-128 | + | 19 | TCAATACTCG TGTGCACCA |
| repeat i4 | 8406-8424 | - | 19 | TCAGCGGACG CAGGGAGGA |
| repeat i3 | 8434-8452 | - | 19 | TCAGGCAACG ACGGGCTGC |
| repeat i2 | 8476-8494 | - | 19 | TCAGAAGCCG ACTGCACTA |
| IRi | 8493-8525 | - | 33 | TGTCGTTTTC AGAAGACGGC TGCACTGAAC GTC |
| repeat i1 | 8499-8517 | - | 19 | TCAGAAGACG GCTGCACTG |
| Name | Associated TE | Coordinates | Orientation | Length | Sequence |
|---|---|---|---|---|---|
| attC qacH_sul1 | In0 | 5857-5890 , 7168-7174 | + | 41 | CCGCTAGCGG GCGGCCGGAA GGTGAATGCT AGGCATCTAA C |
| attC qacH_sul1 core | In0 | 5857-5890 | + | 34 | CCGCTAGCGG GCGGCCGGAA GGTGAATGCT AGGC |
| attI | In0 | 7174-7229 | + | 56 | CTTTGTTTTA GGGCGACTGC CCTGCTGCGT AACATCGTTG CTGCTCCATA ACATCA |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| tniA | In0 | 142-1821 | + | Transposase | |
| tniB delta1 | In0 | 1824-2684 | + | Accessory Gene | |
| istA | IS1326 | 2786-4309 | + | Transposase | None |
| istB | IS1326 | 4296-5081 | + | Accessory Gene | ATPase Transposition Helper |
| GNAT_fam | In0 | 5257-5757 | - | Passenger Gene | Antibiotic Resistance |
| sul1 (ARO:3000410) | In0 | 5885-6724 | - | Passenger Gene | Antibiotic Resistance |
| qacEdelta1 (ARO:3005010) | In0 | 6718-7065 | - | Passenger Gene | Antibiotic Resistance |
| intI1 | In0 | 7310-8323 | + | Integron Integrase | Class 1 |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniA | TniA | In0 | 559 | 142-1821 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | can be extended upstream by 12 amino acids| identical to tniA (Tn1721 and In2)| 25% amino acid sequence identity to TnsB from Tn7 | ||||||||||
| Protein Sequence: | MATDTPRIPE QGVATLPDEA WERARRRAEI ISPLAQSETV GHEAADMAAQ ALGLSRRQVY VLIRRARQGS GLVTDLVPGQ SGGGKGKGRL PEPVERVIHE LLQKRFLTKQ KRSLAAFHRE VTQVCKAQKL RVPARNTVAL RIASLDPRKV IRRREGQDAA RDLQGVGGEP PAVTAPLEQV QIDHTVIDLI VVDDRDRQPI GRPYLTLAID VFTRCVLGMV VTLEAPSAVS VGLCLVHVAC DKRPWLEGLN VEMDWQMSGK PLLLYLDNAA EFKSEALRRG CEQHGIRLDY RPLGQPHYGG IVERIIGTAM QMIHDELPGT TFSNPDQRGD YDSENKAALT LRELERWLTL AVGTYHGSVH NGLLQPPAAR WAEAVARVGV PAVVTRATSF LVDFLPILRR TLTRTGFVID HIHYYADALK PWIARRERWP SFLIRRDPRD ISRIWVLEPE GQHYLEIPYR TLSHPAVTLW EQRQALAKLR QQGREQVDES ALFRMIGQMR EIVTSAQKAT RKARRDADRR QHLKTSARPD KPVPPDTDIA DPQADNLPPA KPFDQIEEW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | tniB delta1 | TniB delta1 | In0 | 286 | 1824-2684 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Function: | probable ATP-binding protein. | ||||||||||
| Comment: | probably truncated by insertion of IS1326::IS1353 | ||||||||||
| Protein Sequence: | MDEYPIIDLS HLLPAAQGLA RLPADERIQR LRADRWIGYP RAVEALNRLE ALYAWPNKQR MPNLLLVGPT NNGKSMIVEK FRRTHPASSD ADQEHIPVLV VQMPSEPSVI RFYVALLAAM GAPLRPRPRL PEMEQLALAL LRKVGVRMLV IDELHNVLAG NSVNRREFLN LLRFLGNELR IPLVGVGTRD AYLAIRSDDQ LENRFEPMML PVWEANDDCC SLLASFAASL PLRRPSPIAT LDMARYLLTR SEGTIGELAH LLMAAAIVAV ESGEEAINHR TLSMAC |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | istA | IstA | IS1326 | 507 | 2786-4309 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Protein Sequence: | MINVAILSAI RRWHFRDGAS IREIARRSGL SRNTVRKYLQ SKVVEPQYPA RDSVGKLSPF EPKLRQWLST EHKKTKKLRR NLRSMYRDLV ALGFTGSYDR VCAFARQWKD SEQFKAQTSG KGCFIPLRFA CGEAFQFDWS EDFARIAGKQ VKLQIAQFKL AHSRAFVLRA YYQQKHEMLF DAHWHAFQIF GGIPKRGIYD NMKTAVDSVG RGKERRVNQR FTAMVSHYLF DAQFCNPASG WEKGQIEKNV QDSRQRLWQG APDFQSLADL NVWLEHRCKA LWSELRHPEL DQTVQEAFAD EQGELMALPN AFDAFVEQTK RVTSTCLVHH EGNRYSVPAS YANRAISLRI YADKLVMAAE GQHIAEHPRL FGSGHARRGH TQYDWHHYLS VLQKKPGALR NGAPFAELPP AFKKLQSILL QRPGGDRDMV EILALVLHHD EGAVLSAVEL ALECGKPSKE HVLNLLGRLT EEPPPKPIPI PKGLRLTLEP QANVNRYDSL RRAHDAA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | istB | IstB | IS1326 | 261 | 4296-5081 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | ATPase Transposition Helper | ||||||||||
| Function: | stimulates transposition | ||||||||||
| Protein Sequence: | MMQHEGHVRI LKSLKLFGMA HAIEELGNQN SPAFNQALPM LDSLIKAEVA EREVRSVNYQ LRVAKFPVYR DLVGFDFSQS LVNEATVKQL HRCDFMEQAQ NVVLIGGPGT GKTHLATAIG TQAVMHLNRR VRFFSTVDLV NALEQEKSSG RQGQIANRLL YADLVILDEL GYLPFSQTGG ALLFHLLSKL YEKTSVILTT NLSFSEWSRV FGDEKMTTAL LDRLTHHCHI LETGNESYRF KHSSTQNKQE EKQTRKLKIE T |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | GNAT_fam | GNAT_fam | In0 | 166 | 5257-5757 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Sequence Family: | Acetyltransf_1 (Pfam:PF00583) | ||||||||||
| Target: | aminoglycoside antibiotic (ARO:0000016) | ||||||||||
| Comment: | putative acetyltransferase ADU64769.1 | ||||||||||
| Protein Sequence: | MDSEEPPNVR VACSGDIDEV VRLMHDAAAW MSAKGTPAWD VARIDRTFAE TFVLRSELLV ASCSDGIVGC CTLSAEDPEF WPDALKGEAA YLHKLAVRRT HAGRGVSSAL IEACRHAART QGCAKLRLDC HPNLRGLYER LGFTHVDTFN PGWDPTFIAE RLELEI |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | sul1 (ARO:3000410) | Sul1 | In0 | 279 | 5885-6724 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic target replacement (ARO:0001002) | ||||||||||
| Sequence Family: | sulfonamide resistant sul (ARO:3004238) | ||||||||||
| Transposase Chemistry: | dihydropteroate synthase | ||||||||||
| Target: | sulfonamide antibiotic (ARO:3000282)||sulfone antibiotic (ARO:3003401) | ||||||||||
| Comment: | perfect match to reference sequence for ARO:3000410 | ||||||||||
| Protein Sequence: | MVTVFGILNL TEDSFFDESR RLDPAGAVTA AIEMLRVGSD VVDVGPAASH PDARPVSPAD EIRRIAPLLD ALSDQMHRVS IDSFQPETQR YALKRGVGYL NDIQGFPDPA LYPDIAEADC RLVVMHSAQR DGIATRTGHL RPEDALDEIV RFFEARVSAL RRSGVAADRL ILDPGMGFFL SPAPETSLHV LSNLQKLKSA LGLPLLVSVS RKSFLGATVG LPVKDLGPAS LAAELHAIGN GADYVRTHAP GDLRSAITFS ETLAKFRSRD ARDRGLDHA |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | qacEdelta1 (ARO:3005010) | QacEdelta1 | In0 | 115 | 6718-7065 | - |
|---|
| Class: | Passenger Gene | ||||||||||
| Subclass: | Antibiotic Resistance | ||||||||||
| Function: | antibiotic efflux (ARO:0010000) | ||||||||||
| Sequence Family: | major facilitator superfamily (MFS) antibiotic efflux pump (ARO:0010002) | ||||||||||
| Target: | disinfecting agents and antiseptics(ARO:3005386) | ||||||||||
| Comment: | subunit of the qac multidrug efflux pump||perfect match to reference sequence for ARO:3005010 (bitscore:219) | ||||||||||
| Protein Sequence: | MKGWLFLVIA IVGEVIATSA LKSSEGFTKL APSAVVIIGY GIAFYFLSLV LKSIPVGVAY AVWSGLGVVI ITAIAWLLHG QKLDAWGFVG MGLIIAAFLL ARSPSWKSLR RPTPW |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | intI1 | IntI1 | In0 | 337 | 7310-8323 | + |
|---|
| Class: | Integron Integrase | ||||||||||
| Subclass: | Class 1 | ||||||||||
| Sequence Family: | Class 1 Integron Tyrosine Integrase | ||||||||||
| Transposase Chemistry: | Tyrosine | ||||||||||
| Protein Sequence: | MKTATAPLPP LRSVKVLDQL RERIRYLHYS LRTEQAYVHW VRAFIRFHGV RHPATLGSSE VEAFLSWLAN ERKVSVSTHR QALAALLFFY GKVLCTDLPW LQEIGRPRPS RRLPVVLTPD EVVRILGFLE GEHRLFAQLL YGTGMRISEG LQLRVKDLDF DHGTIIVREG KGSKDRALML PESLAPSLRE QLSRARAWWL KDQAEGRSGV ALPDALERKY PRAGHSWPWF WVFAQHTHST DPRSGVVRRH HMYDQTFQRA FKRAVEQAGI TKPATPHTLR HSFATALLRS GYDIRTVQDL LGHSDVSTTM IYTHVLKVGG AGVRSPLDAL PPLTSER |
||||||||||
| TnCentral Accession | TE Name | Type | Coordinates | Strand | Length |
|---|---|---|---|---|---|
| IS1326-KY749247.1 | IS1326 | insertion sequence | 2679-5148 | + | 2470 |
| Name | Associated TE | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|---|
| IRL | IS1326 | 2679-2704 | + | 26 | TGTTGAGTTG CATCTAAAAT TGACCC |
| IRR | IS1326 | 5123-5148 | - | 26 | TGTTGATTTG CACCCAAATT TGACCC |
1. Bissonnette L, Roy PH.
Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria.. J Bacteriol. 1992 Feb;174(4):1248-57. doi: 10.1128/jb.174.4.1248-1257.1992. PubMed ID:
1310501
.
2. Recchia GD, Stokes HW, Hall RM. Characterisation of specific and secondary recombination sites recognised by the integron DNA integrase.. Nucleic Acids Res. 1994 Jun 11;22(11):2071-8. doi: 10.1093/nar/22.11.2071. PubMed ID: 8029014 .
3. Hall RM, Brown HJ, Brookes DE, Stokes HW. Integrons found in different locations have identical 5' ends but variable 3' ends.. J Bacteriol. 1994 Oct;176(20):6286-94. doi: 10.1128/jb.176.20.6286-6294.1994. PubMed ID: 7929000 .
4. Brown HJ, Stokes HW, Hall RM. The integrons In0, In2, and In5 are defective transposon derivatives.. J Bacteriol. 1996 Aug;178(15):4429-37. doi: 10.1128/jb.178.15.4429-4437.1996. PubMed ID: 8755869 .
2. Recchia GD, Stokes HW, Hall RM. Characterisation of specific and secondary recombination sites recognised by the integron DNA integrase.. Nucleic Acids Res. 1994 Jun 11;22(11):2071-8. doi: 10.1093/nar/22.11.2071. PubMed ID: 8029014 .
3. Hall RM, Brown HJ, Brookes DE, Stokes HW. Integrons found in different locations have identical 5' ends but variable 3' ends.. J Bacteriol. 1994 Oct;176(20):6286-94. doi: 10.1128/jb.176.20.6286-6294.1994. PubMed ID: 7929000 .
4. Brown HJ, Stokes HW, Hall RM. The integrons In0, In2, and In5 are defective transposon derivatives.. J Bacteriol. 1996 Aug;178(15):4429-37. doi: 10.1128/jb.178.15.4429-4437.1996. PubMed ID: 8755869 .