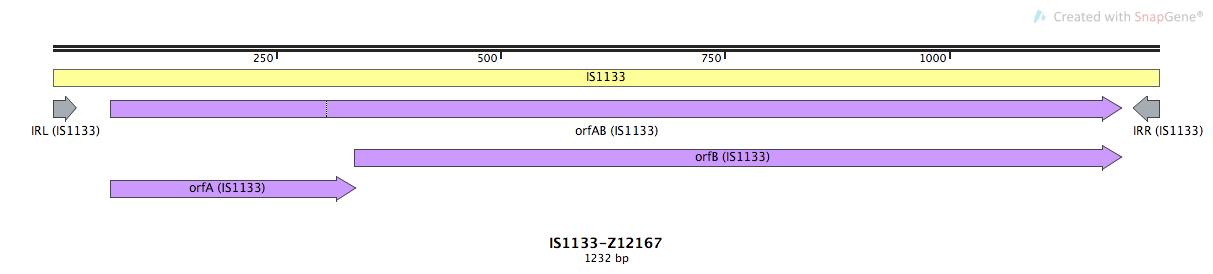
Mobile element type:
Insertion sequence
Name:
IS1133
Synonyms:
Accession:
IS1133-Z12167
Family:
IS3
Group:
IS3
First isolate:
ND
Partial:
ND
Evidence of transposition:
ND
| Name | Coordinates | Direction | Length | Sequence |
|---|---|---|---|---|
| IRL | 1-27 | + | 27 | TGGACTTGCC TCGGTTTCGT TCCGGTC |
| IRR | 1206-1232 | - | 27 | TGGACTTGCA CCGGTTGTGT TCCGGTC |
ORF Summary
| Name | Associated TE | Coordinates | Orientation | Class | Subclass |
|---|---|---|---|---|---|
| orfAB | IS1133 | 65-1194 | + | Transposase | |
| orfA | IS1133 | 65-340 | + | Accessory Gene | Regulator |
| orfB | IS1133 | 337-1194 | + | Transposase |
ORF Details
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | orfAB | OrfAB | IS1133 | 376 | 65-1194 | + |
|---|
| Class: | Transposase | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | fusion protein from -1 programmed frameshifting between orfA and orfB | ||||||||||
| Protein Sequence: | MSLKHSDEFK RDAVRIALTS GLTRRQVASD LSIGLSTLGK WIASISDETK IPTQDTDLLR ENERLRKENR ILREEREILK KAAIFFAVQK L*DFSLLRIT VALSHVHAYV V*WA*QIVVY VHGNAVLHHC ASVVILYF*R IYVSSIGCVW GAMVGRV*QK S*KRWACRLG SVGLDV*CAR ITLQLFERVN SNGQRIVIIP STLHRTY*NK TLAQAHPTRN GQAISLMFGP EKDGSILLLS LTCIPVA*LA GQQVID*SRI LH*GH*IWRW LYANHHRVVF NTQTVGANIA LMNIKSYCSN INCCRP*AGK AIVLITPQ*K ASLNH*RLS* FGADTGKQGE ILRLQSSNI* MAFIIHAEDI QHSAGNRRWH LRKKPL |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | orfA | OrfA | IS1133 | 91 | 65-340 | + |
|---|
| Class: | Accessory Gene | ||||||||||
| Subclass: | Regulator | ||||||||||
| Function: | transposase activity (GO:0004803) | ||||||||||
| Comment: | orfA, together with orfB, encodes a fusion protein formed by translational frame-shifting that is the transposase forIS1133 | ||||||||||
| Protein Sequence: | MSLKHSDEFK RDAVRIALTS GLTRRQVASD LSIGLSTLGK WIASISDETK IPTQDTDLLR ENERLRKENR ILREEREILK KAAIFFAVQK L |
||||||||||
| Gene Name | Protein Name | Associated TE | Gene Length | Coordinates | Strand | orfB | OrfB | IS1133 | 285 | 337-1194 | + |
|---|
| Class: | Transposase | ||||||||||
| Function: | transposase activity (GO:0004803) | ||||||||||
| Transposase Chemistry: | DDE | ||||||||||
| Comment: | orfB, together with orfA, encodes a fusion protein formed by translational frame-shifting that is the transposase for IS1133 | ||||||||||
| Protein Sequence: | MRFQFITDYR GSLSRSRICR LMGVTDRGLR AWKRRPPSLR QRRDLILLAH IREQHRLCLG SYGRPRMTEE LKALGLQVGQ RRVGRLMRQN NITVVRTRKF KRTTDSHHTF NIAPNLLKQD FSASAPNQKW AGDITYVWTR EGWVYLAVIL DLYSRRVIGW ATGDRLKQDL ALRALNMALA LRKPPPGCIQ HTDRGSQYCA HEYQKLLLKH QLLPSMSGKG NCFDNSAVES FFKSLKAELI WRRHWQTRRD IEIAIFEYIN GFYNPRRRHS TLGWKSPVAF EKKAA |
||||||||||
1. Siguier P, Varani A, Perochon J, Chandler M.
Exploring bacterial insertion sequences with ISfinder: objectives, uses, and future developments.. Methods Mol Biol. 2012;859:91-103. doi: 10.1007/978-1-61779-603-6_5. PubMed ID:
22367867
.
2. Heinzel P, Werbitzky O, Distler J, Piepersberg W. A second streptomycin resistance gene from Streptomyces griseus codes for streptomycin-3"-phosphotransferase. Relationships between antibiotic and protein kinases.. Arch Microbiol. 1988;150(2):184-92. doi: 10.1007/BF00425160. PubMed ID: 2844130 .
3. Blot M, Hauer B, Monnet G. The Tn5 bleomycin resistance gene confers improved survival and growth advantage on Escherichia coli.. Mol Gen Genet. 1994 Mar;242(5):595-601. doi: 10.1007/BF00285283. PubMed ID: 7510018 .
4. Yuasa K, Sugiyama M. Bleomycin-induced beta-lactamase overexpression in Escherichia coli carrying a bleomycin-resistance gene from Streptomyces verticillus and its application to screen bleomycin analogues.. FEMS Microbiol Lett. 1995 Oct 1;132(1-2):61-6. doi: 10.1111/j.1574-6968.1995.tb07811.x. PubMed ID: 7590166 .
5. Reznikoff WS. The Tn5 transposon.. Annu Rev Microbiol. 1993;47:945-63. doi: 10.1146/annurev.mi.47.100193.004501. PubMed ID: 7504907 .
6. Steiniger-White M, Rayment I, Reznikoff WS. Structure/function insights into Tn5 transposition.. Curr Opin Struct Biol. 2004 Feb;14(1):50-7. doi: 10.1016/j.sbi.2004.01.008. PubMed ID: 15102449 .
2. Heinzel P, Werbitzky O, Distler J, Piepersberg W. A second streptomycin resistance gene from Streptomyces griseus codes for streptomycin-3"-phosphotransferase. Relationships between antibiotic and protein kinases.. Arch Microbiol. 1988;150(2):184-92. doi: 10.1007/BF00425160. PubMed ID: 2844130 .
3. Blot M, Hauer B, Monnet G. The Tn5 bleomycin resistance gene confers improved survival and growth advantage on Escherichia coli.. Mol Gen Genet. 1994 Mar;242(5):595-601. doi: 10.1007/BF00285283. PubMed ID: 7510018 .
4. Yuasa K, Sugiyama M. Bleomycin-induced beta-lactamase overexpression in Escherichia coli carrying a bleomycin-resistance gene from Streptomyces verticillus and its application to screen bleomycin analogues.. FEMS Microbiol Lett. 1995 Oct 1;132(1-2):61-6. doi: 10.1111/j.1574-6968.1995.tb07811.x. PubMed ID: 7590166 .
5. Reznikoff WS. The Tn5 transposon.. Annu Rev Microbiol. 1993;47:945-63. doi: 10.1146/annurev.mi.47.100193.004501. PubMed ID: 7504907 .
6. Steiniger-White M, Rayment I, Reznikoff WS. Structure/function insights into Tn5 transposition.. Curr Opin Struct Biol. 2004 Feb;14(1):50-7. doi: 10.1016/j.sbi.2004.01.008. PubMed ID: 15102449 .