IS Families/IS6 family
Contents
- 1 General
- 2 Distribution and Phylogenetic Transposase Tree
- 3 Terminal Inverted Repeats.
- 4 Genomic Impact
- 5 Clinical Importance of IS26.
- 6 Organization
- 7 Mechanism: the state of play
- 8 Cointegrate formation
- 9 Circular transposon molecules: translocatable units (TU)
- 10 Targeted transposition.
- 11 Acknowledgements
- 12 Bibliography
General
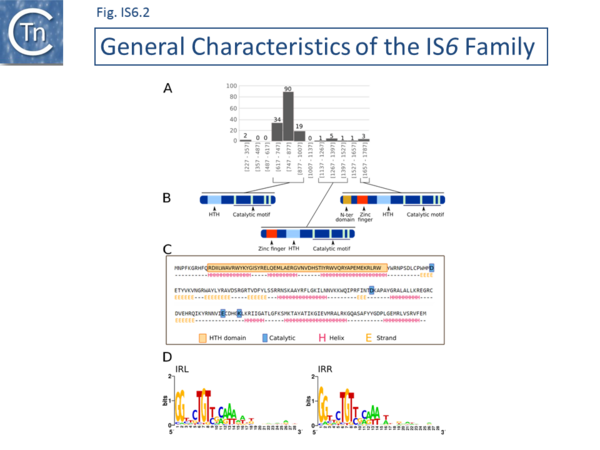
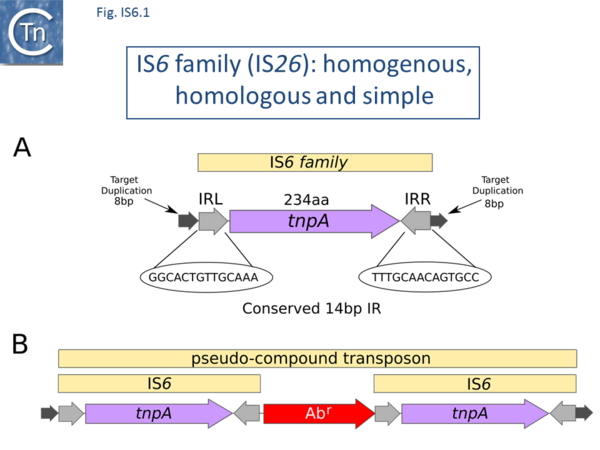
There are at present nearly 160 family members in ISfinder from nearly 80 bacterial and archaeal species but this represents only a fraction of those present in the public databases. The family was named[1] after the directly repeated insertion sequences in transposon Tn6 [2] to standardize the various names that had been attributed to identical elements (e.g. IS15, IS26, IS46, IS140, IS160, IS176) [3][4][5][6][7][8][9][10][11][12][13][14][15], although more recently there has been some attempt to rename the family (see [16]), presumably because of accumulating experimental data from IS26 itself and the importance of this IS in accumulation and transmission of multiple anti biotic resistance, although this might potentially introduce confusion in the literature. IS6 family members have a simple organization (Fig. IS6.1) and generate 8bp direct target repeats on insertion. This family is very homogenous with an average length of about 800 bp and highly conserved short, generally perfect, IRs (Fig. IS6.2). Many are found as part of compound transposons (called pseudo-compound transposons [1] described below) invariably as flanking direct repeats (Fig. IS6.1) a consequence of their transposition mechanism [7][9][13][14][17][18][19][20][21][22][23][24][25][26][27][28][29].
Distribution and Phylogenetic Transposase Tree
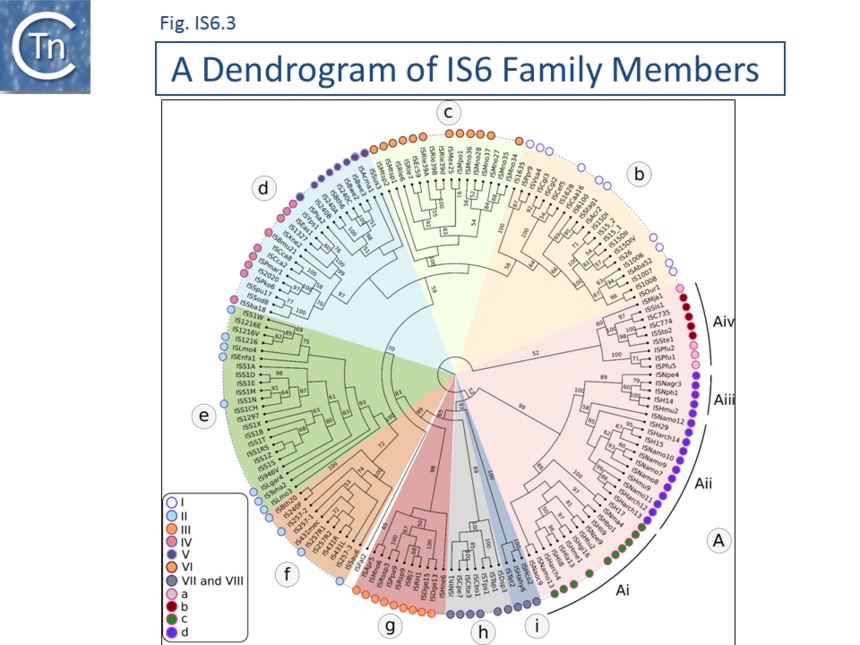
A phylogenetic tree based on the transposase amino acid sequence of the ISfinder collection (Fig. IS6.3) shows that the IS6 family members fall into a number of well-defined clades. This slightly more extensive set of IS corresponds well to the results of another wide-ranging phylogenetic analysis [30]. These clades include one which groups all archaeal IS6 family members (Fig. IS6.3a) composed mainly of Euryarchaeota (Halobacteria ; Fig. IS6.3ai-iii). Group aiv includes both Euryarchaeota (Thermococcales and Methanococcales) and Crenarchaeota (Sulfolobales). Of the 10 clades containing bacterial IS: clade b includes examples from the Alpha-, Beta-, and Gamma-proteobacteria, Firmicutes, Cyanobacteria, Acidobacteria and Bacteroidetes ; clade c is more homogenous and is composed of Alphaproteobacteria (Rhizobiaceae and Methylobacteriaceae); clade d includes some Actinobacteria, Alpha-, Beta-, and Gamma-proteobacteria ; while clades e, f, g and h are composed exclusively of Firmicutes (almost exclusively Lactococci in the case of clades e and f). Clades I and j are more mixed. Clearly, the ISfinder collection does not necessarily reflect the true IS6 family distribution and these grouping should be interpreted with care. For example, although many do not form part of the ISfinderdatabase, IS6 family elements are abundant in archaea and cover almost all of the traditionally recognized archaeal lineages (methanogens, halophiles, thermoacidophiles, and hyperthermophiles [31].
Terminal Inverted Repeats.
The division into clades is also underlined to some extent by the IR sequences. As shown in Fig. IS6.2 (bottom), in spite of the wide range of bacterial and archaeal species in which family members are found, there is a surprising sequence conservation. In particular, the presence of a G dinucleotide at the IS tips and cTGTt and caaa internal motifs. Sequence motifs are more pronounced when each clade is considered separately (Fig. IS6.4).
Clade b
(n=24; with Alpha-, Beta-, and Gamma-proteobacteria, Firmicutes, Cyanobacteria, Acidobacteria and Bacteroidetes) maintains stronger traces of parts of these motifs (GG.. tcTGtt and CAaa).
Clade c
(n= 14; Alphaproteobacteria: Rhizobiaceae and Methylobacteriaceae) shows considerable conservation of an extended motif (GGG... TGTCGCAAA) and some conservation further into both IRL and IRR, although these are different for each end.
Clade d
(n=16; Actinobacteria, Alpha-, Beta-, and Gamma-proteobacteria) includes a well conserved GG..cTGTTGCAAA signature with little conservation further into each end.
Clade e
(n=7) is composed entirely of IS from Lactococcus and, as might be expected, exhibits fully conserved IR (GGTTCTGTTGCAAAGTTT) with significant conservation towards the IS interior, as would be expected in very closely related IS. However, some of the sequence conservation is present in both ends possibly indicating some functional role.
Clade f
(n = 8; Lactococcus and a single Leuconostoc) also exhibits a highly conserved ggTTCTGTTGCAAAGTTT signature at IRL (with only a partially conserved GG at the tip due to just 2 members) which is less conserved in IRR. Again, there is some internal conservation.
Clade g
(n = 8; composed of bacilli: Lysteria, Lactococcus, Enterococcus) also exhibits a similar signature (GGtTctgtTgcaAAgtTt) albeit less conserved. The left and has significant internal conservation.
Clade h
(n = 11; largely Staphylococci with 2 B. thuringiensis) also exhibit the typical GGTTCTGTTGCAAAGTTt signature and some internal conservation in IRL.
Clade i
(n = 1) is more heterogenous (Alpha proteobacteria: Methylobacterium, Paracoccus, Roseovarius, Rhizobium, Bradyrhizobium ; Deinococci and Halobacteria). It contains a poorly conserved IR sequence but does include a prominent gG dinucleotide tip and a poorly pronounced tgtcaagtt signature.
Clade j
Finally, bacterial Clade j (n= 5) composed entirely of Firmicutes (Natranaerobius, Clostridium and Thermoanaerobacter) exhibits a moderately well-defined internal signature TcTgTtAAgTt.
The archaeal-specific clades also generally exhibit well-defined consensus sequences.
Clade Ai
(n = 12) is composed uniquely of Halobacterial Euryarchaeota with a ggtaGTGTTcagatAaG signature and significant internal conservation which is different for each end.
Clade Aii
(n = 5), again, composed entirely of Halobacterial Euryarchaeota (Haloarcula, Halomicrobium, Natronomonas, Natronobacterium, Natrinema) also has well conserved ends, ggtcgTGTTTaGTT, and significant internal conservation which is different for each end.
Clade Aiii
This is also the case for Clade Aiii, also composed of diverse Halobacterial species (Halohasta, Haloferax, Natrinema, Natrialba, Halogeometricum, Natronomonas, Natronococcus, and Haloarcula): GgcACtGTCTAGtT.
Clade Aiv
However, Clade Aiv (n = 9) which includes both Euryarchaeota and Crenarchaeota, has poor conservation although on further analysis, an alignment shows significant conservation in the Sulfolobus and in the Pyrococcus groups with good interior conservation also in the 3 Pyrococcal members. It is possible that the IS ends in the Sulfolobus members have not been accurately identified.
The answer to the recent question:
“An analysis of the IS6/IS26 family of insertion sequences: is it a single family ?”[30] is therefore “Probably, yes”.
Genomic Impact
Activity resulting in horizontal dissemination is suggested, for example, by the observation that copies identical to Mycobacterium fortuitum IS6100 [32](Clade d) occur in other bacteria: as part of a plasmid-associated catabolic transposon carrying genes for nylon degradation in Arthrobacter sp. [33], from the Pseudomonas aeruginosa plasmid R1003 [34], and within the Xanthomonas campestris transposon Tn5393b [35]. Similar copies have also been reported in Salmonella enterica (typhimurium) [36], and on plasmid pACM1 from Klebsiella oxytoca (AF107205) [37].
A single member, ISDsp3, present in single copy in Dehalococcoides sp. BAV1 carries a passenger gene annotated as hypothetical protein.
IS257 [38](Clade h) (also known as IS431) has played an important role in sequestering a variety of antibiotic resistance genes in clinical isolates of methicillin resistant Staphylococcus aureus (MRSA) (e.g.[39][40][41][42]. It provides an outward oriented promoter which drives expression of genes located proximal to the left end. Moreover, both left and right ends appear to carry a –35 promoter component which would permit formation of hybrid promoters on insertion next to a resident –10 element [41][43]. Insertion of can result in activation of a neighboring gene using both a hybrid promoter and an indigenous promoter [41]. IS257 is also involved in expression of tetA [44] and dfrA [42] in S. aureus.
IS26 [6][7][8] (clade d) is encountered with increasing frequency in plasmids of clinical importance where it is involved in expression of antibiotic resistance genes and plasmid rearrangements (see [27][45][46][47][48][49]). Its transposition mechanism contributes to its ability to assemble anti-bacterial resistance genes into clusters (e.g. [50]). It can also form hybrid promoters capable of driving different antibiotic resistance genes: aphA7, blaS2A (Klebsiella pneumoniae [21]), blaSHV-2a (Pseudomonas aeruginosa [51]) and aphA7 (Pasteurella piscicida [52]) as well as the wide spectrum beta-lactam resistance gene blaKPC (Table IS and Gene Expression).
The formation of hybrid promoters on insertion (Table IS and Gene Expression) is clearly a general property of members of the IS6 family [21][41][42][53][54].
Another member, IS6100 [32] (Clade d), often used as an aid in classifying mycobacterial isolates[55] [56][57] has been found to drive strA strB expression in X. campestris pv. vesicatoria, [35].
This IS family is able to form transposons which resemble compound transposons with the flanking IS in direct repeat but, because of the particular transposition mechanism of IS6 family members (see below), were called pseudo-compound transposons [1]. These include Tn610 (flanked by IS6100 [32]), Tn4003 and others (flanked by IS257 [39]) and Tn6023 (flanked by IS26 [58])
Clinical Importance of IS26.
Arrays
Amplification
Cointegrating plasmids.
Organization
Mechanism: the state of play
Cointegrate formation
Circular transposon molecules: translocatable units (TU)
Targeted transposition.
Acknowledgements
We would like to thank Susu He (Nanjing University) for stimulating discussions concerning the transposition models.
Bibliography
- ↑ 1.0 1.1 1.2 Galas DJ, Chandler M. Bacterial Insertion Sequences. In: Berg DE, Howe MM, editors. Mob DNA. Washington, D.C.: American Society for Microbiology; 1989. p. 109–162.
- ↑ Berg DE, Davies J, Allet B, Rochaix JD. Transposition of R factor genes to bacteriophage lambda. ProcNatlAcadSciUSA. 1975;72:3628–3632.
- ↑ Labigne-Roussel A, Courvalin P. IS15, a new insertion sequence widely spread in R plasmids of gram- negative bacteria. MolGenGenet. 1983;189:102–112.
- ↑ Trieu-Cuot P, Courvalin P. Nucleotide sequence of the transposable element IS15. Gene. 1984;30:113–120.
- ↑ <pubmed>2994132</pubmed>
- ↑ 6.0 6.1 <pubmed>PMC326375</pubmed>
- ↑ 7.0 7.1 7.2 <pubmed>PMC326375</pubmed>
- ↑ 8.0 8.1 <pubmed>3003524</pubmed>
- ↑ 9.0 9.1 <pubmed>PMC215669</pubmed>
- ↑ Nucken EJ, Henschke RB, Schmidt FR. Nucleotide-sequence of insertion element IS15 delta IV from plasmid pBP11. DNA Seq. 1990;1:85–88.
- ↑ <pubmed>6304469</pubmed>
- ↑ <pubmed>2999303</pubmed>
- ↑ 13.0 13.1 Colonna B, Bernardini M, Micheli G, Maimone F, Nicoletti M, Casalino M. The Salmonella wien virulence plasmid pZM3 carries Tn1935, a multiresistance transposon containing a composite IS1936- kanamycin resistance element. Plasmid. 1988;20:221–231.
- ↑ 14.0 14.1 <pubmed>PMC162495</pubmed>
- ↑ <pubmed>PMC305975</pubmed>
- ↑ <pubmed>32871211</pubmed>
- ↑ <pubmed>PMC1196216</pubmed>
- ↑ <pubmed>PMC3195058</pubmed>
- ↑ <pubmed>PMC3587239</pubmed>
- ↑ <pubmed>PMC219079</pubmed>
- ↑ 21.0 21.1 21.2 <pubmed>PMC209129</pubmed>
- ↑ Barberis-Maino L, Berger-Bachi B, Weber H, Beck WD, Kayser FH. IS431, a staphylococcal insertion sequence-like element related to IS26 from Proteus vulgaris. Gene. 1987;59:107–113.
- ↑ <pubmed>PMC174916</pubmed>
- ↑ <pubmed>3033719</pubmed>
- ↑ <pubmed>2543009</pubmed>
- ↑ Sundstrom L, Jansson C, Bremer K, Heikkila E, Olsson-Liljequist B, Skold O. A new dhfrVIII trimethoprim-resistance gene, flanked by IS26, whose product is remote from other dihydrofolate reductases in parsimony analysis. Gene. 1995;154:7–14.
- ↑ 27.0 27.1 <pubmed>19074421</pubmed>
- ↑ <pubmed>21393132</pubmed>
- ↑ <pubmed>PMC284528</pubmed>
- ↑ 30.0 30.1 <pubmed>PMC6807381</pubmed>
- ↑ <pubmed>PMC1847376</pubmed>
- ↑ 32.0 32.1 32.2 <pubmed>2163027</pubmed>
- ↑ <pubmed>PMC205175</pubmed>
- ↑ <pubmed>PMC196970</pubmed>
- ↑ 35.0 35.1 <pubmed>PMC167566</pubmed>
- ↑ <pubmed>10930753</pubmed>
- ↑ <pubmed>25291385</pubmed>
- ↑ Rouch D, Skurray R. IS257 from Staphylococcus aureus member of an insertion sequence superfamily Gram-positive and Gram-negative bacteria. Gene. 1989;76:195–205.
- ↑ 39.0 39.1 Rouch DA, Messerotti LJ, Loo SL, Jackson CA, Skurray RA. Trimethoprim resistance transposon Tn4003 from Staphylococcus aureus encodes genes for a dihydrofolate reductase and thymidylate synthetase flanked by three copies of IS257. Mol Microbiol. 1989;3:161–175.
- ↑ Stewart PR, Dubin DT, Chikramane SG, Inglis B, Matthews PR, Poston SM. IS257 and small plasmid insertions in the mec region of the chromosome of Staphylococcus aureus. Plasmid. 1994;31:12–20.
- ↑ 41.0 41.1 41.2 41.3 <pubmed>PMC101884</pubmed>
- ↑ 42.0 42.1 42.2 <pubmed>PMC284724</pubmed>
- ↑ <pubmed>PMC107441</pubmed>
- ↑ <pubmed>7830550</pubmed>
- ↑ <pubmed>PMC1913244</pubmed>
- ↑ <pubmed>23330672</pubmed>
- ↑ <pubmed>16870645</pubmed>
- ↑ <pubmed>23169892</pubmed>
- ↑ <pubmed>20093380</pubmed>
- ↑ <pubmed>PMC4471558</pubmed>
- ↑ <pubmed>PMC89260</pubmed>
- ↑ <pubmed>27873653</pubmed>
- ↑ <pubmed>PMC2443897</pubmed>
- ↑ Allmansberger R, Brau B, Piepersberg W. Genes for gentamicin-(3)-N-acetyl-transferases III and IV. II. Nucleotide sequences of three AAC(3)-III genes and evolutionary aspects. MolGenGenet. 1985;198:514–520.
- ↑ <pubmed>PMC268253</pubmed>
- ↑ <pubmed>PMC330226</pubmed>
- ↑ <pubmed>PMC172007</pubmed>
- ↑ <pubmed>21702681</pubmed>