General Information/Reaction mechanisms
There are a limited number of transposase types defined by their chemistry.
Contents
The main groups
Two groups whose mechanisms are the best understood are the DDE and the HUH superfamilies named after the amino acids which constitute the active sites. The reaction mechanisms involved in transposition using DDE enzymes have been treated in depth (e.g. [1][2][3] as have those of the HUH enzymes (see [4][5]).
Other enzymes
Other types of transposase include: the DEDD enzymes[6] whose chemistry presumably resembles that of the DDE enzymes (since they also have an RNaseH fold[7]) but resemble the RuvC four-way Holliday junction resolvase; Serine transposases which are thought to use chemistry similar to their Serine-site-specific recombinase cousins; and a group of novel transposons, casposons, thought to be primitive ancestors of the CRISPR system[8].
The transpososome
Most transposition reactions can be divided into several defined steps generally comprising: binding of the transposase to the ends; elaboration of a synaptic complex involving the transposase, perhaps accessory proteins, and both transposon ends - this step involves either concomitant or subsequent (depending on the element) recruitment of the target DNA; cleavage and strand transfer of the transposon ends into the target; and processing of the strand transfer complex to a final product. The protein-DNA complexes assembled during this process are called transpososomes.
Enzymes or reaction components?
An important point which has not yet been fully addressed is whether transposases are true enzymes which can be recycled or whether they are themselves reaction components which are consumed during the process. The fact that many transposases show a preference for action on the element from which they are expressed (in cis) and the coupling between transposase translation and DNA binding[9] (10) imposes strong constraints on transposase recycling. Moreover, at least in the case of bacteriophage Mu, special machinery in place for removing transposase from the transposition complex following insertion which includes ClpX[10][11][12]. In addition, the Lon protease is implicated in proteolysis of the IS903 transposase[13][14].
Chemistry of the DDE enzymes
The DDE catalytic site
It has become clear that many of the enzymes involved in transposition reactions are related and, moreover, are part of a larger family of phosphoryltransferases which also includes RNaseH and the RuvC "Holliday junction resolvase"[15][16][17][18][19][20][21][22].
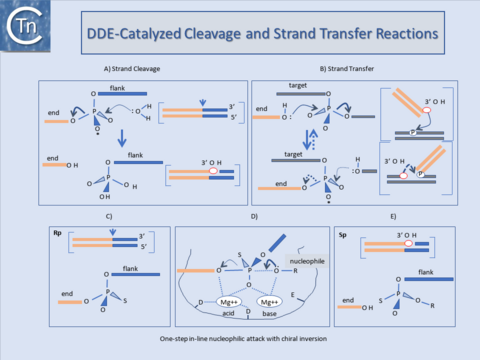
These transposases catalyse cleavage at the 3' ends of the element by an attacking nucleophile (generally H2O) to expose a free 3'OH group (Fig.1.40.1). This hydroxyl in turn acts as a nucleophile to attack a 5' phosphate group in the target DNA (strand transfer) in a single-step transesterification reaction. Under certain conditions the enzyme is also capable of "disintegrating" the transposon end by catalysing the attack of the 3' target OH group on the new transposon-target junction[23][24][25].
The reaction(s) do not require an external energy source and do not involve a covalently linked enzyme-substrate intermediate as do certain site-specific recombination reactions. It is worth underlining that, since it is the donor strand itself which performs the cleavage-ligation step in the target DNA, no cleaved target molecule is detected in the absence of strand transfer.
An acidic amino acid triad (DDE : Asp, Asp, Glu) present in these enzymes is intimately involved in catalysis presumably by co-ordinating divalent metal cations (in particular Mg2+) which assist the various nucleophilic attacking- and leaving-groups during the course of the reaction (Fig.1.40.1). The reaction is an in-line nucleophilic attack resulting in chiral inversion of the target phosphate. Chiral inversion has been observed for retroviral integration[26][27], bacteriophage Mu[28], and Tn10[29] transposition and in a related reaction, V(D)J recombination[30] and was revealed by substituting a non-bridging oxygen for a sulphur group which fixes the normally achiral phosphate group in one or other of its alternative chiral forms (Fig.1.40.1). For many ISs and the retroviral integrases (IN) this triad is known as the DD(35)E motif and is highly conserved[31][32][33][34]. In addition, alignments of several Tpases[35] revealed regions of amino acid conservation designated, N1, N2, and N3 and C1[36] which encompass the D (N2), D (N3) and E (C1) regions of typical DDE motifs respectively. The C1 region is probably the most defined structural element. It appears to be part of an a-helix with additional conserved amino acids including a K or R residue approximately 7 amino acids or two helical turns downstream from the E residue[37][38][39]. Less well-conserved residues often occur at approximately one helical turn (3 or 4 residues) upstream and downstream E (DDE motifs). For retroviruses this has been shown to interact with the terminal base pairs of the element presumably contributing to correct positioning of the transposon end in the active site[40][41].
Although this conservation in the primary sequence is lower in certain of the other groups of elements and not all families have been explored in sufficient detail to assure that the alignments are biologically relevant, mutagenesis studies with some of these elements (e.g. the Mu, Tn7, IS10, and Tc1/3 Tpases and the retroviral integrases) clearly underline the importance of these residues. Moreover, structural analysis showed that these acidic amino acids are arranged close to each other in a similar, three-dimensional manner in other phosphoryltransferases such as RNaseH and RuvC[42][43], otherwise unrelated to Tpases. These primary amino acid conservations are also reflected in conserved structural features. Major conserved features identified in the structures of the catalytic cores of retroviral integrases (Fig.1.40.1), and the Mu and IS50 Tpases include 2 beta-sheets each harboring one of the D residues and the long beta-helix including the E residue. The alfa-helix is designated α-4 in HIV and C1 in the Tpases[44][45], see [46]). It is one of the most conserved regions in the catalytic core. Mutagenesis and crosslinking studies with INHIV suggested that it played a role in positioning both the nucleophile and viral DNA[47][48][49]. In particular K159 or K156, which cross-link to the terminal CA dinucleotide, are located on the same side of the alfa-helix and are strongly conserved. Q148 also lies on the same side and appears to interact with the terminal end of the non-processed strand (see [50]).
An amide or basic amino acid is highly conserved at this or the neighboring position and mutation results in severe impairment of catalysis (IN: [51]; IS10: [52]; IS50: see [53]; IS903: [54].
Transposition reactions and different types of gene rearrangement.
If DDE transposases are capable of catalysing only single strand cleavage to generate a 3’OH at the end of the transposon, how do IS elements with move from one place to another While initiation of a transposition reaction catalysed by the DDE transposases proceeds via transfer of the 3' end of the transposon (transferred strand), the outcome of the reaction is governed by cleavage of the 5' (non-transferred) end of the element as discussed in Groups with DDE Transposases (Fig.1.8.2).
Cleavage of the transferred strand alone=
5' strand cleavage does not occur concomitantly with 3' strand transfer (Fig.1.8.2), the donor and target molecules become covalently linked. Subsequent 5' strand cleavage will separate the element from the donor backbone and will also result in direct insertion (Fig.1.8.2). In the case of retroviruses, only the 3' cleavage occurs, removing 2 bp from the end of the double strand DNA viral copy. However, since no donor backbone is attached to the viral DNA, direct insertion can ensue. In cases where the 5’ transposon end remains attached to the donor backbone DNA the result of 3' strand transfer is to join transposon and target leaving a 3'OH in the target DNA at the junction. This can act as a primer for replication of the element and generate cointegrates where donor and target molecules are separated by a single transposon copy at each junction. During its lytic cycle bacteriophage Mu similarly undergoes only 3' cleavage of the transferred strand, the donor backbone remains attached and cointegrate molecules result if replication occurs. Members of the Tn3 family appear to transpose in this way and certain Tn7 and IS903 mutants can be induced to undergo cointegrate formation.
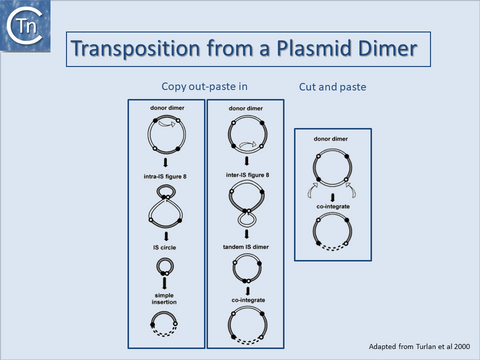
Cointegrates from donor dimer plasmids
It is important to note that cointegrates identical to those produced by replicative transposition can also be produced by a non-replicative process either from a plasmid dimer[55][56][57] (Fig.1.40.2) or from tandemly repeated copies of an IS element[58][59][60][61][62][63]. For copy out – paste in transposition, the two outcomes lead to either simple insertion ((Fig.1.40.2, left) or cointegrates (Fig.1.40.2, middle). For cut and paste transposition, a dimer plasmid donor gives rise directly to a cointegrate structure (Fig.1.40.2, right).
Transposase structures and the Transpososome
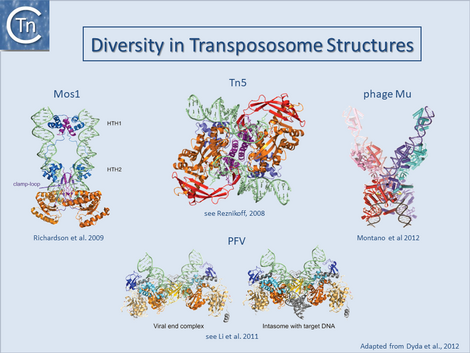
Only a single prokaryotic IS DDE transposase, that of IS50, has yielded structural information[64][65] and this has provided important insights into transposition of its cognate IS. The only other prokaryotic DDE whose structure is available is that of the transpososome of bacteriophage Mu[66]. On the other hand, a limited number of transposase and DDE transposase-DNA co-complex structures are available and have yielded important insights into the details of the transposition reaction (Fig.1.40.3) and (Fig.1.8.3).
They all tend to support the detailed reaction mechanism. These include several retroviral integrases (IN) such as HIV, ASV and PFV[67][68][69][70][71][72][73][74]; the mariner transposon Mos1[75][76][77][78]; and the hAT transposon HERMES (revealing a complex set of interactions between neighboring monomers in the octomeric transposase complex[79][80][81].
These studies underline a large degree of diversity in the different transposase/integrase and in their architecture and the detailed interaction with their cognate DNA sequences even though they share a well conserved catalytic domain topology. A recurring theme in all these structures is that the complexes tend to be stabilised by a network of inter- and intra-protein contacts and contacts with the DNA. Moreover, the cleavages appear to occur “in trans”: cleavages on one end are catalysed by a transposase molecule bound to the other DNA end[82][83][84].
This phenomenon includes both prokaryotic transposable elements Tn5[85] and Mu[86][87] as well as the eukaryotic Mos1 transposon[88] and retroviruses[89][90]. This arrangement represents an important regulatory mechanism since no chemistry can occur on a single isolated transposon end which would result in non-productive transposition and would have a negative impact on the host replicon.
Cleavage can only be initiated in the context of an architecturally defined and properly assembled “transpososome” in which both DNA ends are synapsed and which can give rise to productive transposition.
Chemistry of HUH enzymes
Mechanism and Overall Protein Architecture
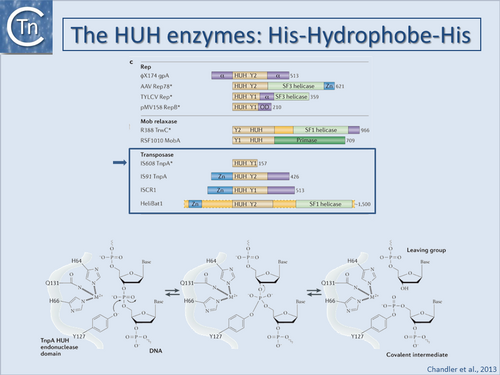
Historically, bacteriophage Φ-X174 protein A (gpA) was the first identified HUH superfamily member although, surprisingly, no structural information is available. Many related proteins subsequently identified by bioinformatics[91][92] included proteins involved in catalysis of viral and plasmid rolling circle replication (RCR), conjugative plasmid transfer, and DNA transposition[93][94][95][96][97]. They all carry conserved protein motifs, including the "HUH" motif composed of two histidine (H) residues separated by a bulky hydrophobic (U) residue, and the Y-motif containing either one or two tyrosine (Tyr) residues (found in Y1 and Y2 enzymes respectively).
Y1 HUH enzymes (Fig.1.41.1) include Rep proteins of some plasmids with ssDNA replication intermediates (such as pUB110[98]), a wide range of eukaryotic viruses[99], most conjugative plasmid relaxases[100][101], ISCR (insertion sequences related to IS91[102]) and IS200/IS605 insertion sequence family transposases[103][104]. Y2 enzymes include ΦX174 gpA itself, Rep proteins of other isometric ssDNA and dsDNA phages (e.g. phage P2[105]), some cyanobacterial and archaeal plasmids and parvoviruses (e.g. adeno-associated virus, AAV) as well as transposases of the IS91 and helitron families[106], and MOBF family plasmid relaxases. In some cases, both Y residues are mechanistically important while for others, only one of the pair appears to be essential.
HUH enzymes use a unique mechanism for catalysing ssDNA breakage and joining. The active site tyrosine creates a 5'-phosphotyrosine intermediate and a free 3'-OH at the cleavage site (Fig.1.41.1). The 3'-OH can be used for different tasks. The most obvious is to prime replication, as observed for HUH domains in single-stranded phage Rep proteins, RCR plasmids and conjugative relaxases. The 3'-OH group can also act as the nucleophile for strand transfer to resolve the phosphotyrosine intermediate in the termination step of RCR replication, conjugative transfer and transposition. The HUH enzyme cleavage polarity is inverse to that of the tyrosine recombinases, which make 3' phosphotyrosine intermediates and generate free 5'-OH groups that cannot be used as replication primers[107]. HUH enzymes also require a divalent metal ion to facilitate cleavage by localizing and polarising the scissile phosphodiester bond in contrast to the cofactor-independent tyrosine recombinases. Depending on the enzyme, Mg2+, Mn2+ or other divalent metal ions can be used in vitro[108][109][110][111][112][113]. It is presumed that Mg2+ or Mn2+ are the physiological cofactors. The HUH histidine pair provides two of the three ligands necessary for metal ion coordination (Fig.1.41.1). The location and identity of the third, invariably polar (Glu, Asp, His or Gln) residue varies across the superfamily.
3D structures of several Rep and relaxase HUH domains with and without bound DNA are available [e.g. [114][115][116][117][118][119][120]]. The order of HUH and Y motifs varies in the primary sequence: in the Relaxase group, the Y-motif is upstream of the HUH-motif whereas in the Rep group it is downstream (Fig.1.41.1). This “circular permutation” [121][122][123] changes the domain topology. Nevertheless, the three-dimensional constellation of active site residues is virtually identical across the superfamily.
Given the diverse HUH protein functions, it is not surprising that other domains are often appended to the HUH domain (Fig.1.41.1). These are often of unknown function but, ATP dependent helicase, zinc binding, primase and multimerisation domains are recurring themes. For example, the ssDNA substrates needed by HUH enzymes can be generated by a dedicated DNA helicase domain C-terminal to the HUH domain[124][125][126][127] or alternatively by recruitment of a host helicase. RCR processes use 3´-5´helicase activity acting on the template strand to facilitate DNA unwinding at the replication fork while in conjugation, helicases (as part of the relaxase) are transported into the recipient cell and track 5´to 3´on the transported ssDNA.
DNA Recognition
Many HUH nucleases recognize and bind DNA hairpin structures with cleavage sites located within the hairpin or in the ssDNA on the 5' or 3' side of the stem. The crucial role of hairpins has been firmly established in many systems including plasmid conjugation, eukaryotic viral and plasmid replication and transposition[128][129][130][131][132][133][134]. In other systems, palindromic sequences that can form DNA hairpins are present near the probable HUH nuclease cleavage sites [135]. Such hairpins can be formed in vivo under a number of physiological conditions (see [136]).
Structural studies revealed that small DNA hairpins can be recognized in several different ways: sequence-specific recognition of the dsDNA stem; structure-specific recognition of irregularities in the stem; or sequence-specific recognition of the hairpin loop[137][138][139][140][141][142].
The hairpin-flanking DNA - in many cases in single-stranded form - is also often important for recognition. Relaxases, for example, make extensive contacts with the bases extending between the hairpin and cleavage site[143][144][145], and for a relative of IS200/IS605 transposases, TnpAREP, nucleotides on the 5' side of the hairpin are crucial for binding and sequence-specific recognition[146]. Other family members[147][148] [23,39], have more complex binding modes.
HUH enzymes as transposases
Transposases of members of the IS200/IS605[149], IS91[150] and ISCR[151] insertion sequence families and the eukaryotic helitrons[152] are also HUH enzymes. Those of the IS200/IS605 family are the best understood.
IS200/IS605 family
IS200/IS605 family transposases are single domain proteins with only the essential HUH motif and a single catalytic Tyr (Y1 transposases, (Fig.1.41.1)). Both TnpAIS608 and TnpAISDra2 are obligatory dimers and the active sites are believed to adopt two functionally important conformations, one in which each is composed of the HUH motif from one monomer and the Tyr residue carried by an alpha-helix (αD) from the other (trans configuration), and the other in which both motifs are contributed by the same monomer (cis configuration). Only the former has been observed crystallographically. Similar proteins are sometimes found associated with repeated Extragenic Palindromes (REP sequences whose hairpin structures resemble the ends of IS200/IS605 family members.
RCR transposons: IS91, ISCR and Helitron families
The earliest identified HUH domain transposases were those of the IS91 family and are significantly larger than Y1 transposases (Fig.1.41.1), carry a Y2 motif and include an N-terminal zinc binding motif and additional domains of yet unidentified function. A group of related elements, the ISCRs often associated with a variety of antibiotic resistance genes (see [153]) carry an orf (the CR or common region) resembling IS91 family transposases but with only a single Tyr (Fig.1.41.1). In addition, eukaryotic relatives, the Helitrons, have been identified by bioinformatic approaches.
Chemistry of DEDD enzymes
DEDD enzymes are limited to transposases of the IS110 family at present[154] and closely related to the Piv and MooV invertases from Moraxella lacunata / M. bovis[155][156] and N. gonorrhoeae[157][158] respectively. Piv catalyses inversion of a DNA segment permitting expression of a type IV pilin. However, the organization of IS110 family members and the inversion systems are different. In the IS, the recombinase is located within the element whereas in the inversion systems it is located outside the invertible segment[159].
Although it has proved difficult to determine the activity of these transposases in detail in vitro, transposition of ISs with DEDD Tpases may be unusual and involve HJ intermediates which must be resolved using a RuvC-like mechanism. This type of recombination would be consistent with the close relationship between DEDD transposases and the Piv/MooV invertases which presumably resolve HJ structures during inversion[160].
Chemistry of Serine-transposases
Other TE encode transposases which resemble serine site-specific recombinases. Their chemistry is presumably similar to that of their sequence-specific recombinase cousins. IS607 family members encode a Tpase closely resembling serine site-specific recombinases which use serine as a nucleophile for cleavage of the DNA strand. At present little is known about transposition of this IS family although it is thought that these elements generate circular intermediates (NDF Grindley pers. comm. cited in [161]). Presumably the enzyme catalyses similar cleavages and strand transfers as its site-specific serine recombinase cousins using a transitory 5’ phosphoserine covalent intermediate. Based on transposase structures from structural genomics studies and detailed knowledge of the general serine recombinase mechanism, [162] have proposed a model for the transposition mechanism. This includes a synaptic transposase tetramer (as for classical serine recombinases). The model explains the lack of target specificity exhibited in IS607 transposition[163], behavior which is unusual for this type of recombinase.
Bibliography
- ↑ <pubmed>1383220</pubmed>
- ↑ <pubmed>9112436</pubmed>
- ↑ <pubmed>26104718</pubmed>
- ↑ <pubmed>23832240</pubmed>
- ↑ <pubmed>26350330</pubmed>
- ↑ <pubmed>15866929</pubmed>
- ↑ <pubmed>7923356</pubmed>
- ↑ <pubmed>24884953</pubmed>
- ↑ <pubmed>22195971</pubmed>
- ↑ <pubmed>8022280</pubmed>
- ↑ <pubmed>8631314</pubmed>
- ↑ <pubmed>7557391</pubmed>
- ↑ <pubmed>2161528</pubmed>
- ↑ <pubmed>8898394</pubmed>
- ↑ <pubmed>8696976</pubmed>
- ↑ <pubmed>7628012</pubmed>
- ↑ <pubmed>21439812</pubmed>
- ↑ <pubmed>23135398</pubmed>
- ↑ <pubmed>7801124</pubmed>
- ↑ <pubmed>20067338</pubmed>
- ↑ <pubmed>23217365</pubmed>
- ↑ <pubmed>26104718</pubmed>
- ↑ <pubmed>1738845</pubmed>
- ↑ <pubmed>8950268</pubmed>
- ↑ <pubmed>8391505</pubmed>
- ↑ <pubmed>1760846</pubmed>
- ↑ <pubmed>10559232</pubmed>
- ↑ <pubmed>1649006</pubmed>
- ↑ <pubmed>10847684</pubmed>
- ↑ <pubmed>8599117</pubmed>
- ↑ <pubmed>1963920</pubmed>
- ↑ <pubmed>1314954</pubmed>
- ↑ <pubmed>1647013</pubmed>
- ↑ <pubmed>21518873</pubmed>
- ↑ <pubmed>7934941</pubmed>
- ↑ <pubmed>7934941</pubmed>
- ↑ <pubmed>7752887</pubmed>
- ↑ <pubmed>9362498</pubmed>
- ↑ <pubmed>8302872</pubmed>
- ↑ <pubmed>9362498</pubmed>
- ↑ <pubmed>9755183</pubmed>
- ↑ <pubmed>8696976</pubmed>
- ↑ <pubmed>7628012</pubmed>
- ↑ <pubmed>7934941</pubmed>
- ↑ <pubmed>9689049</pubmed>
- ↑ <pubmed>10547692</pubmed>
- ↑ <pubmed>9362498</pubmed>
- ↑ <pubmed>9755183</pubmed>
- ↑ <pubmed>9573274</pubmed>
- ↑ <pubmed>26104441</pubmed>
- ↑ <pubmed>9573274</pubmed>
- ↑ <pubmed>8565068</pubmed>
- ↑ <pubmed>18680433</pubmed>
- ↑ <pubmed>9417930</pubmed>
- ↑ <pubmed>6298776</pubmed>
- ↑ <pubmed>2832702</pubmed>
- ↑ <pubmed>6298776</pubmed>
- ↑ <pubmed>10540284</pubmed>
- ↑ <pubmed>9393723</pubmed>
- ↑ <pubmed>3034717</pubmed>
- ↑ <pubmed>3146616</pubmed>
- ↑ <pubmed>1645443</pubmed>
- ↑ <pubmed>10760133</pubmed>
- ↑ <pubmed>10207011</pubmed>
- ↑ <pubmed>10884228</pubmed>
- ↑ <pubmed>23135398</pubmed>
- ↑ <pubmed>26104441</pubmed>
- ↑ <pubmed>19036793</pubmed>
- ↑ <pubmed>19609359</pubmed>
- ↑ <pubmed>19132083</pubmed>
- ↑ <pubmed>21216426</pubmed>
- ↑ <pubmed>21277766</pubmed>
- ↑ <pubmed>20428106</pubmed>
- ↑ <pubmed>26061770</pubmed>
- ↑ <pubmed>15103153</pubmed>
- ↑ <pubmed>17565190</pubmed>
- ↑ <pubmed>19766564</pubmed>
- ↑ <pubmed>23262225</pubmed>
- ↑ <pubmed>16511103</pubmed>
- ↑ <pubmed>16041385</pubmed>
- ↑ <pubmed>25036632</pubmed>
- ↑ <pubmed>21439812</pubmed>
- ↑ <pubmed>20067338</pubmed>
- ↑ <pubmed>23217365</pubmed>
- ↑ <pubmed>10908658</pubmed>
- ↑ <pubmed>8612279</pubmed>
- ↑ <pubmed>8612278</pubmed>
- ↑ <pubmed>25662605</pubmed>
- ↑ <pubmed>21068843</pubmed>
- ↑ <pubmed>20118915</pubmed>
- ↑ <pubmed>1630899</pubmed>
- ↑ <pubmed>8374079</pubmed>
- ↑ <pubmed>11447285</pubmed>
- ↑ <pubmed>16163392</pubmed>
- ↑ <pubmed>16209952</pubmed>
- ↑ <pubmed>16760305</pubmed>
- ↑ <pubmed>19396961</pubmed>
- ↑ <pubmed>2666843</pubmed>
- ↑ <pubmed>22760663</pubmed>
- ↑ <pubmed>19919603</pubmed>
- ↑ <pubmed>21876676</pubmed>
- ↑ <pubmed>16760305</pubmed>
- ↑ <pubmed>16163392</pubmed>
- ↑ <pubmed>16209952</pubmed>
- ↑ <pubmed>11327759</pubmed>
- ↑ <pubmed>11447285</pubmed>
- ↑ <pubmed>16756503</pubmed>
- ↑ <pubmed>20890269</pubmed>
- ↑ <pubmed>16540117</pubmed>
- ↑ <pubmed>19440202</pubmed>
- ↑ <pubmed>14604527</pubmed>
- ↑ <pubmed>16216584</pubmed>
- ↑ <pubmed>23359708</pubmed>
- ↑ <pubmed>16540117</pubmed>
- ↑ <pubmed>19440202</pubmed>
- ↑ <pubmed>14604527</pubmed>
- ↑ <pubmed>22885300</pubmed>
- ↑ <pubmed>14967147</pubmed>
- ↑ <pubmed>14625590</pubmed>
- ↑ <pubmed>12876370</pubmed>
- ↑ <pubmed>8374079</pubmed>
- ↑ <pubmed>14625590</pubmed>
- ↑ <pubmed>14604517</pubmed>
- ↑ <pubmed>11447285</pubmed>
- ↑ <pubmed>16943286</pubmed>
- ↑ <pubmed>10516041</pubmed>
- ↑ <pubmed>2159383</pubmed>
- ↑ <pubmed>16163392</pubmed>
- ↑ <pubmed>16209952</pubmed>
- ↑ <pubmed>16540117</pubmed>
- ↑ <pubmed>22885300</pubmed>
- ↑ <pubmed>10933682</pubmed>
- ↑ <pubmed>22199259</pubmed>
- ↑ <pubmed>8523519</pubmed>
- ↑ <pubmed>9618448</pubmed>
- ↑ <pubmed>21119018</pubmed>
- ↑ <pubmed>16209952</pubmed>
- ↑ <pubmed>21119018</pubmed>
- ↑ <pubmed>23359708</pubmed>
- ↑ <pubmed>22885300</pubmed>
- ↑ <pubmed>14967147</pubmed>
- ↑ <pubmed>14625590</pubmed>
- ↑ <pubmed>23359708</pubmed>
- ↑ <pubmed>14625590</pubmed>
- ↑ <pubmed>12876370</pubmed>
- ↑ <pubmed>22885300</pubmed>
- ↑ <pubmed>14967147</pubmed>
- ↑ <pubmed>17267412</pubmed>
- ↑ <pubmed>16163392</pubmed>
- ↑ <pubmed>8127907</pubmed>
- ↑ <pubmed>16760305</pubmed>
- ↑ <pubmed>11447285</pubmed>
- ↑ <pubmed>16760305</pubmed>
- ↑ <pubmed>11169105</pubmed>
- ↑ <pubmed>2403542</pubmed>
- ↑ <pubmed>9079926</pubmed>
- ↑ <pubmed>15687191</pubmed>
- ↑ <pubmed>12897009</pubmed>
- ↑ <pubmed>15866929</pubmed>
- ↑ <pubmed>10092658</pubmed>
- ↑ <pubmed>17347521</pubmed>
- ↑ <pubmed>24195768</pubmed>
- ↑ <pubmed>10986230</pubmed>