Difference between revisions of "General Information/What Is an IS?"
| Line 1: | Line 1: | ||
==Classical IS== | ==Classical IS== | ||
| − | The original definition of an IS [[:Image:1.3.1.png|(Fig | + | The original definition of an IS [[:Image:1.3.1.png|(Fig.3.1)]] was: a short, generally phenotypically cryptic, DNA segment encoding only the enzymes necessary for its transposition and capable of repeated insertion into many different sites within a genome using mechanisms independent of large regions of DNA homology between the IS and target <ref><nowiki><pubmed>26104715</pubmed></nowiki></ref>. Classical IS are between 0.7 and 2.5 kb in length, genetically compact with one or two open reading frames (orfs) which occupy the entire length of the IS and terminate in flanking imperfect terminal repeat sequences (IR) (<b>Table 1</b>). The orfs include the Tpase that catalyzes the DNA cleavages and strand transfers leading to IS movement and, in some cases, regulatory proteins. Their highly compact nature is illustrated by the fact that some IS have developed “recoding” strategies such as [[wikipedia:Ribosomal_frameshift|Programmed Ribosomal Frameshifting]] (involving ribosome slippage) and Programmed Transcriptional Realignment (involving RNA polymerase [[wikipedia:Slipped_strand_mispairing|slippage]]) <ref><nowiki><pubmed> 21673094</pubmed></nowiki></ref><ref><nowiki><pubmed>24499397</pubmed></nowiki></ref><ref><nowiki><pubmed>26350305</pubmed></nowiki></ref><ref><nowiki><pubmed> 21478364</pubmed></nowiki></ref><ref><nowiki><pubmed>11125107</pubmed></nowiki></ref><ref><nowiki><pubmed>8384687</pubmed></nowiki></ref><ref><nowiki><pubmed>12762024</pubmed></nowiki></ref>. |
These permit assembly of different functional protein domains effectively encoding two proteins of different functions in one DNA segment. IS also often generates a short flanking directly repeated duplication (DR) of the target DNA on insertion. These characteristics are not limited to prokaryotic IS but are also shared with most eukaryotic DNA transposons. Classical IS generally transpose using a double-strand DNA intermediate. | These permit assembly of different functional protein domains effectively encoding two proteins of different functions in one DNA segment. IS also often generates a short flanking directly repeated duplication (DR) of the target DNA on insertion. These characteristics are not limited to prokaryotic IS but are also shared with most eukaryotic DNA transposons. Classical IS generally transpose using a double-strand DNA intermediate. | ||
However, for prokaryotic IS, this strict definition has been broadened over the years with the discovery of an increasing number of non-canonical derivatives and variants, some of which are described in the following sections. Moreover, as we learn more about diversity from sequenced genomes, classification is becoming more problematic because the large degree of MGE diversity is obscuring the borders between certain types of TE ([[General Information/Fuzzy Borders|Fuzzy Borders]]) <ref><nowiki><pubmed>24499397</pubmed></nowiki></ref>. Despite their abundance and diversity, the number of different chemical mechanisms used in TE movement is surprisingly limited and many quite divergent TE share a similar mechanism. | However, for prokaryotic IS, this strict definition has been broadened over the years with the discovery of an increasing number of non-canonical derivatives and variants, some of which are described in the following sections. Moreover, as we learn more about diversity from sequenced genomes, classification is becoming more problematic because the large degree of MGE diversity is obscuring the borders between certain types of TE ([[General Information/Fuzzy Borders|Fuzzy Borders]]) <ref><nowiki><pubmed>24499397</pubmed></nowiki></ref>. Despite their abundance and diversity, the number of different chemical mechanisms used in TE movement is surprisingly limited and many quite divergent TE share a similar mechanism. | ||
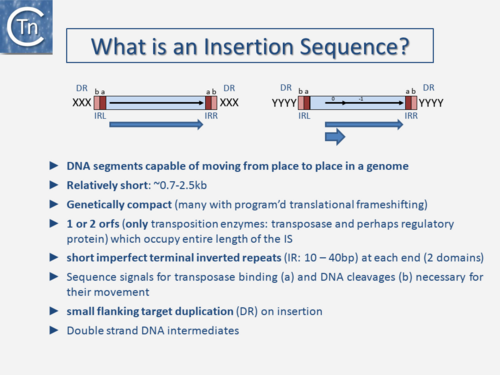
| − | [[File:1.3.1.png|link=https://tncentral.ncc.unesp.br/TnPedia/index.php/File:1.3.1.png|alt=|border|center|thumb|500x500px|'''Fig | + | [[File:1.3.1.png|link=https://tncentral.ncc.unesp.br/TnPedia/index.php/File:1.3.1.png|alt=|border|center|thumb|500x500px|'''Fig.3.1.''' What is an Insertion Sequence? Terminal inverted repeats (IRL and IRR) are shown as two-colored boxes (a and b) with functions for transposase binding (a) and recognition for cleavage and strand transfer (a). A single (left) or double (right) open reading frame is shown underneath the IS (blue arrow). The transposase of the IS on the right is produced by programmed -1 translational frameshifting. The reading frames are indicated within the IS. The product of the upstream frame generally acts as a regulatory protein. The indigenous Tpase promoter is shown located (by convention) in IRL. XXX and YYYY represent the short direct target repeat sequence which is generally duplicated during the insertion event.]] |
==Characteristics of insertion sequence families== | ==Characteristics of insertion sequence families== | ||
| Line 171: | Line 171: | ||
==New types of IS== | ==New types of IS== | ||
| − | '''<big>O</big>'''ne example of this expanding diversity is the identification of another entire class of IS <ref><nowiki><pubmed>10986230</pubmed></nowiki></ref><ref><nowiki><pubmed>9858724</pubmed></nowiki></ref><ref><nowiki><pubmed>11807059</pubmed></nowiki></ref>. Members of this class use an entirely different mechanism of transposition involving single-strand circular DNA intermediates which appear to target stalled replication forks <ref><nowiki><pubmed>26350330</pubmed></nowiki></ref> [[:Image:1.3.2.png|(Fig | + | '''<big>O</big>'''ne example of this expanding diversity is the identification of another entire class of IS <ref><nowiki><pubmed>10986230</pubmed></nowiki></ref><ref><nowiki><pubmed>9858724</pubmed></nowiki></ref><ref><nowiki><pubmed>11807059</pubmed></nowiki></ref>. Members of this class use an entirely different mechanism of transposition involving single-strand circular DNA intermediates which appear to target stalled replication forks <ref><nowiki><pubmed>26350330</pubmed></nowiki></ref> [[:Image:1.3.2.png|(Fig.3.2)]]. They possess small transposases (~150 aa) which are completely different to the classical IS in the type of chemistry they catalyze ([[General Information/Major Groups are Defined by the Type of Transposase They Use#Groups with HUH Enzymes|Groups with HUH Enzymes]]). |
| − | Another example are the [[General Information/The casposases|casposons]] which are related to CRISPRs but whose transposition has yet to be fully characterized <ref><nowiki><pubmed>24884953</pubmed></nowiki></ref><ref><nowiki><pubmed>28472712</pubmed></nowiki></ref><ref><nowiki><pubmed>28683354</pubmed></nowiki></ref><ref><nowiki><pubmed>26104718</pubmed></nowiki></ref>. | + | Another example are the [[General Information/The casposases|casposons]] which are related to [[wikipedia:CRISPR|CRISPRs]] but whose transposition has yet to be fully characterized <ref><nowiki><pubmed>24884953</pubmed></nowiki></ref><ref><nowiki><pubmed>28472712</pubmed></nowiki></ref><ref><nowiki><pubmed>28683354</pubmed></nowiki></ref><ref><nowiki><pubmed>26104718</pubmed></nowiki></ref>. |
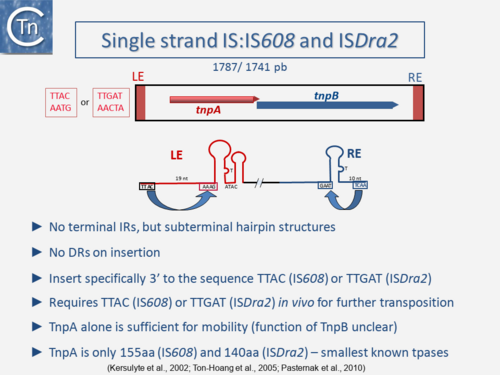
| − | [[Image:1.3.2.png|thumb|500x500px|'''Fig | + | [[Image:1.3.2.png|thumb|500x500px|'''Fig.3.2.''' Organization of the IS''200''/IS''605'' family insertion sequences which transpose using single-strand DNA intermediates. The top of the figure shows the genetic organization. The generic IS is shown as a box. The element-specific tetra- (IS''608'') or penta- (IS''Dra2'') nucleotide target site is shown boxed in red on the left. Left (LE) and right (RE) ends carrying subterminal hairpins are presented as red boxes. The transposase reading frame, ''tnpA'', is shown within the IS as a red line and the accessory frame ''tnpB'' as a blue line. The direction of transcription is indicated by the arrowhead. |
The bottom of the figure shows the subterminal secondary structures at the left (red) end LE and the right (blue) end RE. The left and right cleavage sites are presented as black | The bottom of the figure shows the subterminal secondary structures at the left (red) end LE and the right (blue) end RE. The left and right cleavage sites are presented as black | ||
Revision as of 17:27, 21 May 2020
Contents
Classical IS
The original definition of an IS (Fig.3.1) was: a short, generally phenotypically cryptic, DNA segment encoding only the enzymes necessary for its transposition and capable of repeated insertion into many different sites within a genome using mechanisms independent of large regions of DNA homology between the IS and target [1]. Classical IS are between 0.7 and 2.5 kb in length, genetically compact with one or two open reading frames (orfs) which occupy the entire length of the IS and terminate in flanking imperfect terminal repeat sequences (IR) (Table 1). The orfs include the Tpase that catalyzes the DNA cleavages and strand transfers leading to IS movement and, in some cases, regulatory proteins. Their highly compact nature is illustrated by the fact that some IS have developed “recoding” strategies such as Programmed Ribosomal Frameshifting (involving ribosome slippage) and Programmed Transcriptional Realignment (involving RNA polymerase slippage) [2][3][4][5][6][7][8].
These permit assembly of different functional protein domains effectively encoding two proteins of different functions in one DNA segment. IS also often generates a short flanking directly repeated duplication (DR) of the target DNA on insertion. These characteristics are not limited to prokaryotic IS but are also shared with most eukaryotic DNA transposons. Classical IS generally transpose using a double-strand DNA intermediate. However, for prokaryotic IS, this strict definition has been broadened over the years with the discovery of an increasing number of non-canonical derivatives and variants, some of which are described in the following sections. Moreover, as we learn more about diversity from sequenced genomes, classification is becoming more problematic because the large degree of MGE diversity is obscuring the borders between certain types of TE (Fuzzy Borders) [9]. Despite their abundance and diversity, the number of different chemical mechanisms used in TE movement is surprisingly limited and many quite divergent TE share a similar mechanism.
Characteristics of insertion sequence families
| Table 1. Abbreviations: DR, duplication repeat; IS, insertion sequence; ORF, open reading frame. | |||||||||
| Families | Sub-Groups | Typical size-range | DR (bp) | Ends | IRs | No ORFs | Frameshift | Catalytic residues | Mechanism |
|---|---|---|---|---|---|---|---|---|---|
| IS1 | — | 740–1180 | 8–9 | GGnnnTG | Y | 2 | ORFAB | DDE | copy-and-paste and cointegrate |
| single ORF | 800–1200 | 0–9 | N | 1 | — | ||||
| ISMhu11 | 900–4600 | 0–10 | Y | 2 | ORFAB | ||||
| IS1595 | ISPna2 | 1000–1150 | 8 | GGCnnTG | Y | 1 | — | DDNK | copy-and-paste? |
| ISPna2+pass | 1500–2600 | 8 | — | — | |||||
| ISH4 | 1000 | 8 | CGCTCTT | DDNK | |||||
| IS1016 | 700–745 | 7–9 | GGGgctg | DDEK | |||||
| IS1595 | 900–1100 | 8 | CcTGATT | DDNK+ER4R7 | |||||
| ISSod11 | 1000–1100 | 8 | nnnGcnTATC | DDHK+ER4R7 | |||||
| ISNwi1 | 1080–1200 | 8 | ggnnatTAT | DDEK+ER4 | |||||
| ISNwi1+pass | 1750–4750 | 8 | — | — | |||||
| ISNha5 | 3450–7900 | 8 | CGGnnTT | DDER/K | |||||
| IS3 | IS150 | 1200–1600 | 3–4 | TG | Y | 2 | ORFAB | DDE | copy-and-paste |
| IS407 | 1100–1400 | 4 | TG | ||||||
| IS51 | 1000–1400 | 3–4 | TG | ||||||
| IS3 | 1150–1750 | 3–4 | TGa/g | ||||||
| IS2 | 1300–1400 | 5 | TG | ||||||
| IS481 | 950–1300 | 4–15 | TGT | Y | 1 | — | DDE | copy-paste? | |
| ISNCY | IS1202 | 1400–1700 | 5 | TGT | Y | 1 | — | DDEQ | — |
| IS4 | IS10 | 1200–1350 | 9 | CT | Y | 1 | DDE | hairpin intermediate | cut-and-paste |
| IS50 | 1350–1550 | 8–9 | C | hairpin intermediate | |||||
| ISPepr1 | 1500–1600 | 7–8 | -T-AA | ? | |||||
| IS4 | 1400–1600 | 10–13 | -AAT | ? | |||||
| IS4Sa | 1150–1750 | 8–10 | CA | ? | |||||
| ISH8 | 1400–1800 | 10 | ? | ||||||
| IS231 | 1450–5400 | 10–12 | CAT | 1 or + * | *passenger genes | ||||
| IS701 | — | 1400–1550 | 4 | — | Y | 1 | — | DDE | — |
| ISAba11 | — | — | |||||||
| ISH3 | — | 1225–1500 | 4–5 | C-GT | Y | 1 | — | DDE | — |
| IS1634 | — | 1500–2000 | 5–6 | C | Y | 1 | — | DDE | — |
| IS5 | IS903 | 950–1150 | 9 | GG | Y | 1 | — | DDE | — |
| ISL2 | 850–1200 | 2–3 | — | ||||||
| ISH1 | 900–1150 | 8 | -GC | ||||||
| IS5 | 1000–1500 | 4 | Ga/g | ||||||
| IS1031 | 850–1050 | 3 | GAa/g | ||||||
| IS427 | 800–1000 | 2–4 | Ga/g | 2 | ORFAB | ||||
| IS1182 | — | 1330–1950 | 0–60 | — | Y | 1 | — | DDE | — |
| ISNCY | ISDol1 | 1600–1900 | 6–7 | — | Y | 1 | — | DDE | — |
| IS6 | — | 700–900 | 8 | GG | Y | 1 | — | DDE | co-integrate |
| IS21 | — | 1750–2600 | 4–8 | TG | Y | 2 * | — | DDE | — |
| IS30 | — | 1000–1700 | 2–3 | — | Y | 1 | — | DDE | copy-and-paste |
| IS66 | — | 2000–3000 | 8–9 | GTAA | Y | 3* | — | DDE* | — |
| ISBst12 | 1350–1900 | 1 | DDE | ||||||
| IS256 | — | 1200–1500 | 8–9 | Ga/g | Y | 1 | — | DDE | copy-and-paste |
| IS1249 | 1300 | 0–10 | GG | ||||||
| ISC1250 | 1250 | 0–9 | GG | ||||||
| ISH6 | — | 1450 | 8 | GGT | Y | 1 | — | DDE | — |
| ISLre2 | — | 1500–2000 | 9 | — | Y | 1 | — | DDE | — |
| ISKra4 | ISAzba1 | 1400–2900 | 0 | — | Y | 1 or + * | — | DDE | — |
| ISMich2 | 1250–1400 | 8 | GGG | 1 or 2 | ORFAB | ||||
| ISKra4 | 1400–3700 | 9 | GGG | 1 or + * | — | ||||
| IS630 | — | 1000–1400 | 2* | — | Y | 1 or 2 | ORFAB | DDE | cut-and-paste |
| IS982 | — | 1000 | 3–9 | AC | Y | 1 | — | DDE | — |
| IS1380 | — | 1550–2000 | 4–5 | CC | Y | 1 | — | DDE | — |
| ISAs1 | — | 1200–1500 | 8–10 | CAGGG | Y | 1 | — | — | — |
| ISL3 | — | 1300–2300 | 8 | GG | Y | 1 | — | — | — |
| Tn3 | — | >3000 | 0 | GGGG | Y | >1 | — | DDE | co-integrate |
| ISAzo13 | — | 1250–2200 | 0–4 | Ga/g | Y | 1 | — | — | — |
| IS110 | — | 1200–1550 | 0 | — | N | 1 | — | DEDD | — |
| IS1111 | — | — | — | Y* | — | — | — | — | |
| IS91 | — | 1500–2000 | 0 | — | N | 1 | — | HUH/Y2 | rolling circle |
| IS200/IS605 | IS200 | 600–750 | 0 | — | 0 | 1* | — | HUH/Y1 | peel-and-paste |
| IS605 | 1300–2000 | — | — | — | 2* | — | HUH/Y1** | ||
| IS607 | — | 1700–2500 | 0 | — | N | 2* | — | Serine** | — |
| ISNCY | IS892 | 1600 | 0–8 | CTAG | Y | 2 | ORFAB | — | — |
| ISLbi1 | 1400–1500 | 5 | — | Y | 1 | ||||
| ISMae2 | 1400–2400 | 9 | CAG | Y | 1 | ||||
| ISPlu15 | 800–1000 | 0 | — | N | 1 | ||||
| ISA1214 | 1000–1200 | 8–12 | — | Y | 2 | ||||
| ISC1217 | 1200 | 6–8 | TAG | Y | 1 | ||||
| ISM1 | 1300–1600 | 8–9 | — | Y | 1 | ||||
New types of IS
One example of this expanding diversity is the identification of another entire class of IS [10][11][12]. Members of this class use an entirely different mechanism of transposition involving single-strand circular DNA intermediates which appear to target stalled replication forks [13] (Fig.3.2). They possess small transposases (~150 aa) which are completely different to the classical IS in the type of chemistry they catalyze (Groups with HUH Enzymes). Another example are the casposons which are related to CRISPRs but whose transposition has yet to be fully characterized [14][15][16][17].
Bibliography
- ↑ <pubmed>26104715</pubmed>
- ↑ <pubmed> 21673094</pubmed>
- ↑ <pubmed>24499397</pubmed>
- ↑ <pubmed>26350305</pubmed>
- ↑ <pubmed> 21478364</pubmed>
- ↑ <pubmed>11125107</pubmed>
- ↑ <pubmed>8384687</pubmed>
- ↑ <pubmed>12762024</pubmed>
- ↑ <pubmed>24499397</pubmed>
- ↑ <pubmed>10986230</pubmed>
- ↑ <pubmed>9858724</pubmed>
- ↑ <pubmed>11807059</pubmed>
- ↑ <pubmed>26350330</pubmed>
- ↑ <pubmed>24884953</pubmed>
- ↑ <pubmed>28472712</pubmed>
- ↑ <pubmed>28683354</pubmed>
- ↑ <pubmed>26104718</pubmed>