IS Families/IS66 family
Contents
General
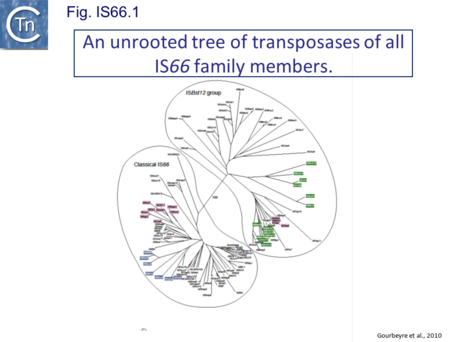
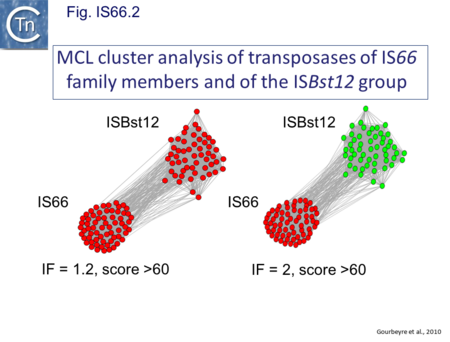
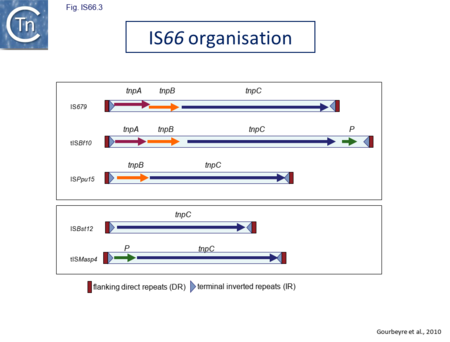
IS66 was first identified in the Ti plasmid pTi66 of Agrobacterium tumefaciens Sciaky[1][2] and, soon after in the symbiotic plasmid, pSym, of Rhizobium fredi[3]. The vast majority of IS66 members originate from the Proteobacteria with several from the Bacteriodetes/Chlorobi and the Firmicutes. A second group of closely related ISs, widely spread among both bacteria and archaea are thought to represent a sub-group within the IS66 family[4]. These are relatively well distributed (Fig. IS66.1). The founder member, ISBst12, originally isolated from Bacillus stearothermophilus, was described as a novel family[5], but identification of many additional examples suggests that the ISBst12 and IS66 groups should be considered a single family (Fig. IS66.2). Several examples of IS derivatives with passenger genes have been identified (Fig. IS66.3; Table IS66.1). One important example is a potential tIS in which IRL is located downstream from an MCR-3 (colistin resistance) gene[6]). Members of the ISBst12 group are found in Actinobacteria, Cyanobacteria, Deinococcus/Thermus, Firmicutes, and Planctomycetes as well as in Proteobacteria. They are also found in the Euryarchaeota phylum of archaea (but have not yet been identified in the Crenarchaeota).
Genome Impact
Members of this family are involved in insertional gene inactivation (e.g. inactivation of the liaFSR operon leading to hypersensitivity to daptomycin[9]; loss of loss of S-layer-gene expression in Bacillus stearothermophilus[10]; gene disruption in the cyanobacterium Fremyella duplosiphon involved in regulation of phycoerythrin synthesis[11] and in the gnaA (encoding UDP-N-acetylglucosamine C-6 dehydrogenase) of Acenitobacter baumannii leading to changes in the antibacterial resistance profile[12]).
It has also been shown (using RACE) that IS66 insertion can create hybrid promoters[13].
Organization
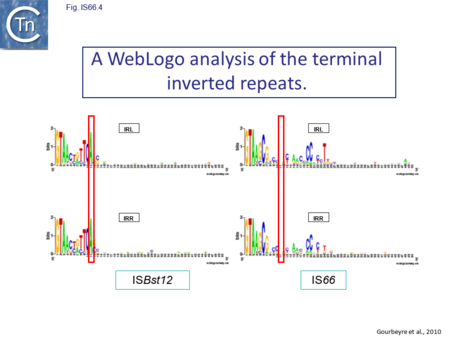
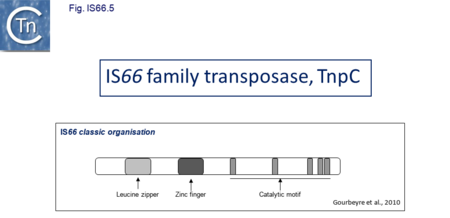
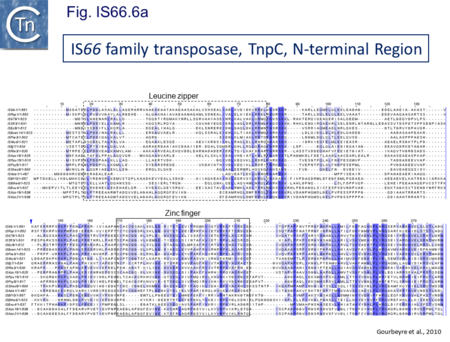
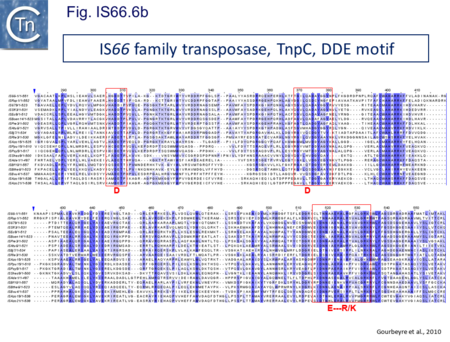
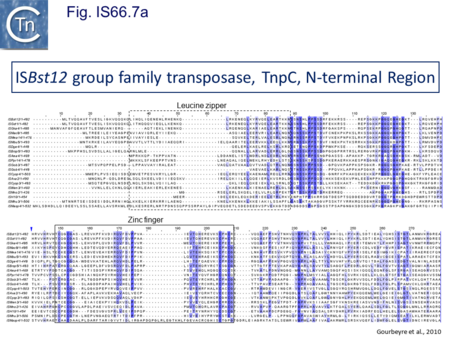
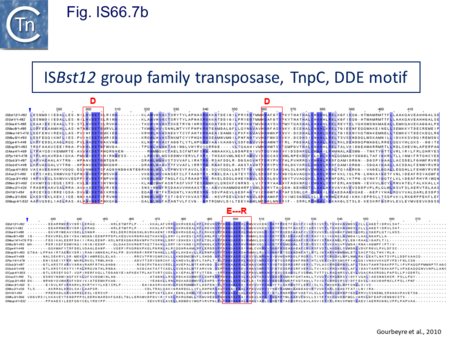
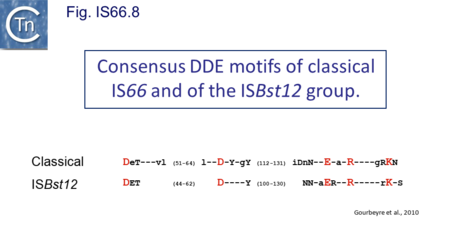
The IS66 reference copy from a plasmid of the enteropathogenic Escherichia coli B171, IS679[14] is defined by three orfs (Fig. IS66.3): tnpA, tnpB and tnpC and relatively well conserved terminal IR of about 20-30 bp flanked by an 8 bp DR at their insertion sites (Fig. IS66.4). Orf tnpC (Fig. IS66.5) is 1572 bp and its predicted product includes an N-terminal region with potential leucine zipper and zinc finger motifs (Fig. IS66.5; Fig. IS66.6a; Fig. IS66.7a) and a typical DDE motif (Fig. IS66.5; Fig. IS66.6b; Fig. IS66.7b; outlined in Fig. IS66.8). It also carries an insertion domain between the second D and the E of the DDE motif (e.g.IS679, ISPsy5 and ISMac8)[15] (see Fig.1.8.3) (Table Transposases examined by secondary structure prediction programs).
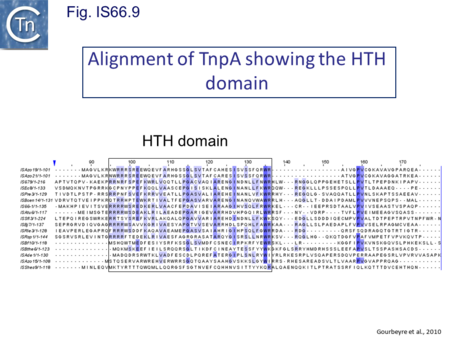
The role of the products of tnpA (651 bp) and tnpB (345 bp) is less clear. TnpA carries a potential HTH motif (Fig. IS66.9) while TnpB shows no marked potential secondary structure motifs. Mutation of each orf separately (by introduction of an in-frame deletion) reduced transposition by at least two orders of magnitude[16]. The three frames are disposed in a pattern suggesting translational coupling: tnpB is in general in translational reading frame -1 compared to tnpA and in most cases the termination codon of tnpA and the initiation codon of tnpB overlap (ATGA). An initiation codon for tnpC occurs slightly downstream separated from tnpB by about 20 bp.
However, rather surprisingly, in the light of a requirement for all three orfs for transposition of the canonical IS66 family member IS679, members of the ISBst12 group are devoid of tnpA and tnpB and carry only the tnpC reading frame. Although both ISBst12 and IS66 members contain IRs which start with 5’GTAA3’, they are clearly distinguishable due to a single conserved A at bp 11 In ISBst12 which is not conserved in IS66 (Fig. IS66.4; Table IS66.1).
IS66 members can be grouped into three classes based on their organization: those including all three orfs, A, B and C transcribed in the same direction; those with additional passenger genes invariably present downstream of orfC and transcribed in the same direction; and those which lack orfA but retain both orfs B and C (Fig. IS66.3; Table IS66.1). Each of these organizations includes members with multiple copies, implying that they are active in transposition. In addition to the DDE catalytic domain (Fig. IS66.5; Fig. IS66.6b; Fig. IS66.7b)[17], TnpC also exhibits a highly conserved CwAH-rR motif downstream of the second D residue, a relatively conserved CX2(C)X33CX2C motif characteristic of a zinc finger (ZF) further upstream and a leucine-rich region which might form a leucine zipper (LZ) necessary in multimerisation of other Tpases [18], at the N-terminus (Fig. IS66.5; Fig. IS66.6a; Fig. IS66.7a).
A list of representative IS66 family members and the ISBst12 group
| Table IS66.1. A list of representative IS66 family members and the ISBst12 group. The table summarises from left to right: the IS name; group defined in the MCL analysis; accession number; host from which it was identified; kingdom (archaea, A, or bacteria, B); phylum; organisation (org) A,B,C,P show the presence of TnpA, TnpB, TnpC and Passenger genes respectively; length of the IS in base pairs (bp); length of the terminal inverted repeats (IR); length of flanking direct repeats (DR); number of examples identified in the host genome. | ||||||||||
| IS Name | Group | Accession number | Host | A/B | G+/G- | Org. | L (bp) | IR (bp) | DR (bp) | N° |
|---|---|---|---|---|---|---|---|---|---|---|
| IS66-1 | — | AF242881 | Agrobacterium tumefaciens | B | G- | ABC | 2556 | 18/20 | 8 | 2 |
| IS679 | — | NC_002142 | Escherichia coli | B | G- | ABC | 2704 | 17/25 | 8 | 6 |
| ISAde1 | — | NC_011891 | Anaeromyxobacter dehalogenans 2CP-1 | B | G- | ABC | 2957 | 20/27 | 8 | 2 |
| ISAtu6 | — | NC_010929 | Agrobacterium tumefaciens | B | G- | ABCP | 2798 | 23/24 | 8 | 3 |
| ISAzo15 | — | NC_006513 | Azoarcus sp. EbN1 or Aromatoleum aromaticum EbN1 | B | G- | ABC | 2441 | 20/21 | 8 | 4 |
| ISAzo19 | — | NC_006513 | Azoarcus sp. EbN1 or Aromatoleum aromaticum EbN1 | B | G- | ABC | 2423 | 23/26 | 8 | 3 |
| ISAzo21 | — | NC_006513 | Azoarcus sp. EbN1 or Aromatoleum aromaticum EbN1 | B | G- | ABC | 2423 | 23/26 | 8 | 2 |
| ISBcen14 | — | NC_011001 | Burkholderia cenocepacia J2315 | B | G- | ABC | 2516 | 18/22 | 8 | 4 |
| ISBf10 | — | NC_006347 | Bacteroides fragilis YCH46 | B | G- | ABCP | 2939 | 20/21 | 8 | 4 |
| ISBj7 | — | NC_004463 | Bradyrhizobium japonicum USDA 110 | B | G- | ABC | 2865 | 40/50 | 8 | 2 |
| ISBthe6 | — | NC_004663 | Bacteroides thetaiotaomicron | B | G- | ABC | 2544 | 24 | 8 | 2 |
| ISBvu4 | — | NC_009614 | Bacteroides vulgatus ATCC 8482 | B | G- | BC | 2371 | 30/32 | 8 | 5 |
| ISEc8 | — | NC_004431 | Escherichia coli CFT073 | B | G- | ABC | 2442 | 18/22 | 8 | 7 |
| ISPpu15 | — | NC_002947 | Pseudomonas putida KT2440 | B | G- | BC | 2041 | 22/28 | 8 | 4 |
| ISPre3 | — | NC_004444 | Pseudomonas resinovorans | B | G- | ABCP | 2957 | 17/24 | 8 | 2 |
| ISPsy5 | — | AE016853 | Pseudomonas syringae pv. tomato str. DC3000 | B | G- | BC | 2059 | 21/28 | 8 | 33 |
| ISRle3 | — | NC_008382 | Rhizobium leguminosarum bv. viciae 3841 | B | G- | ABC | 2500 | 14/15 | 8 | 2 |
| ISRsp1 | — | U00090 | Rhizobium sp. NGR234 | B | G- | ABCP | 3481 | 17/22 | 8 | 2 |
| ISSfl3 | — | AL391753 | Shigella flexneri | B | G- | ABC | 2729 | 11 | 0 | 2 |
| ISShes9 | — | NC_008750 | Shewanella sp. W3-18-1 | B | G- | ABC | 2370 | 18/24 | 8 | 3 |
| ISAma4 | ISBst12 | NC_011138 | Alteromonas macleodii 'Deep ecotype' | B | G- | C | 1529 | 28/29 | 9 | 3 |
| ISAva2 | ISBst12 | NC_007410 | Anabaena variabilis ATCC 29413 | B | G- | C | 1547 | 14 | 8 | 9 |
| ISBrsp5 | ISBst12 | NC_009445 | Bradyrhizobium sp. ORS278 | B | G- | C | 1541 | 18/19 | 8 | 2 |
| ISBst12 | ISBst12 | AF162268 | Bacillus stearothermophilus | B | G+ | C | 1612 | 15/16 | 8 | 15 |
| ISCysp3 | ISBst12 | NZ_AAXW00000000 | Cyanothece sp. CCY 0110 | B | G- | C | 1522 | 14 | 8 | 18 |
| ISCysp4 | ISBst12 | NC_011884 | Cyanothece sp. PCC 7425 | B | G- | C | 1585 | 14/17 | 8 | 12 |
| ISDge4 | ISBst12 | NC_008025 | Deinococcus geothermalis DSM 11300 | B | G+ | C | 1453 | 18/25 | 8 | 6 |
| ISGka4 | ISBst12 | NC_006509 | Geobacillus kaustophilus HTA426 | B | G+ | C | 1635 | 21/24 | 8 | 3 |
| ISGob3 | ISBst12 | NZ_ABGO00000000 | Gemmata obscuriglobus UQM 2246 | B | G- | C | 1597 | 11/12 | 8 | 8 |
| ISGob4 | ISBst12 | NZ_ABGO00000000 | Gemmata obscuriglobus UQM 2246 | B | G- | C | 1576 | 14/15 | 8 | 6 |
| ISGst1 | ISBst12 | NC_010420 | Geobacillus stearothermophilus | B | G+ | C | 1613 | 18/26 | 8 | 3 |
| ISH10 | ISBst12 | NC_002607 | Halobacterium sp. NRC-1 | A | — | C | 1584 | 16/18 | 8 | 5 |
| ISMac8 | ISBst12 | NC_003552 | Methanosarcina acetivorans C2A | A | — | C | 1603 | 13/15 | 8 | 3 |
| ISMasp4 | ISBst12 | NC_008576 | Magnetococcus sp. MC-1 | B | G- | PC | 1969 | 19 | 8 | 7 |
| ISMbu5 | ISBst12 | NC_007955 | Methanococcoides burtonii DSM 6242 | A | — | C | 1696 | 18/19 | 8/0 | 9 |
| ISMhu3 | ISBst12 | NC_007796 | Methanospirillum hungatei JF-1 | A | — | PC | 1727 | 9/12 | 0 | 2 |
| ISMma14 | ISBst12 | NC_003901 | Methanosarcina mazei Go1 | A | — | C | 1529 | 22/30 | 8 | 9 |
| ISMno2 | ISBst12 | NC_011894 | Methylobacterium nodulans ORS 2060 | B | G- | C | 1419 | 21/27 | 8 | 8 |
| ISPpr14 | ISBst12 | NC_006371 | Photobacterium profundum SS9 | B | G- | C | 1568 | 19/27 | 8/9 | 9 |
| ISWen3 | ISBst12 | NZ_AAGB00000000 | Wolbachia endosymbiont of Drosophila ananassae | B | G- | C | 1482 | 18/20 | 8-0 | 30 |
Mechanism and Insertion Specificity
Nothing is known about the transposition mechanism of this group of IS and they exhibit no substantial target sequence specificity.
Bibliography
- ↑ <pubmed>6095299</pubmed>
- ↑ <pubmed>6366736</pubmed>
- ↑ <pubmed>24302303</pubmed>
- ↑ <pubmed>20079432</pubmed>
- ↑ <pubmed>10974105</pubmed>
- ↑ <pubmed>29712655</pubmed>
- ↑ <pubmed>1633570</pubmed>
- ↑ <pubmed>18792942</pubmed>
- ↑ <pubmed>27353469</pubmed>
- ↑ <pubmed>10974105</pubmed>
- ↑ <pubmed>21888899</pubmed>
- ↑ <pubmed>31358579</pubmed>
- ↑ <pubmed>29374029</pubmed>
- ↑ <pubmed>11418571</pubmed>
- ↑ <pubmed>20067338</pubmed>
- ↑ <pubmed>11418571</pubmed>
- ↑ <pubmed>20079432</pubmed>
- ↑ <pubmed>9761671</pubmed>