General Information/Influence of transposition mechanisms on genome impact
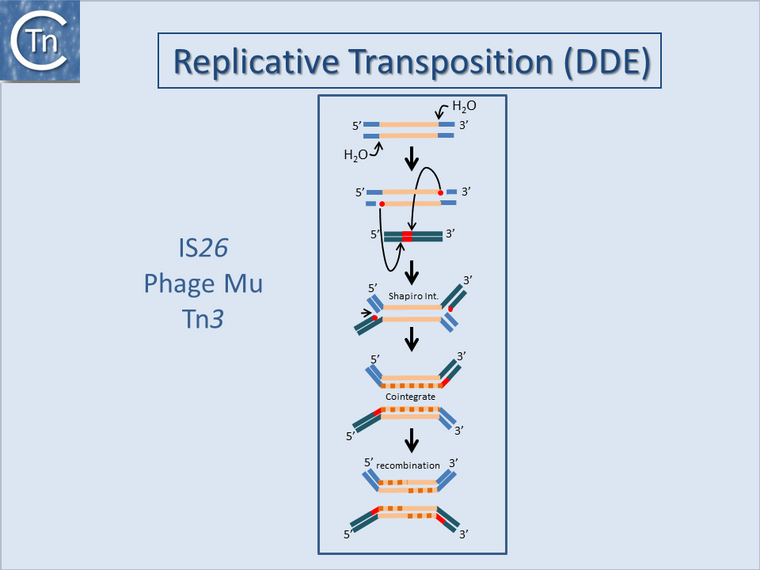
The way in which strand cleavages and transfers occur during transposition also affects the outcome of the transposition events and therefore impinges on genome structure. For IS with DDE Tpases, Tn3 and IS6 family members generate fusions or cointegrates between the donor and target replicons by a process of replicative transposition, presumably by Target Primed Replicative Transposition (TPRT)[1][2][3] (Fig.17.1 and Fig.17.2 top). However, in the event of intramolecular transposition, this type of mechanism is expected to give rise to inversions with a copy of the IS at each junction or inversions with a single IS copy remaining and a second copy segregating with a circularized deletion[4] (Fig.17.2 botton; see [5]). Note that similar effects are also known to occur by homologous recombination between two inverted or directly repeated IS copies in a replicon. Other known mechanisms such as cut-and-paste, or copy-and-paste (Donor Primed Replicatice Transposition; DPRT)[6] would not generate this type of genomic rearrangement but could contribute to genomic modifications in other ways such as “nearly precise excision”[7] or by using alternative sequences which resemble their IR[8][9].

Bibliography
- ↑ Hickman AB, Chandler M, Dyda F . Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. - Crit Rev Biochem Mol Biol: 2010 Feb, 45(1);50-69 [PubMed:20067338] [DOI]
- ↑ Hickman AB, Dyda F . Mechanisms of DNA Transposition. - Microbiol Spectr: 2015 Apr, 3(2);MDNA3-0034-2014 [PubMed:26104718] [DOI]
- ↑ Siguier P, Gourbeyre E, Varani A, Ton-Hoang B, Chandler M . Everyman's Guide to Bacterial Insertion Sequences. - Microbiol Spectr: 2015 Apr, 3(2);MDNA3-0030-2014 [PubMed:26104715] [DOI]
- ↑ Shapiro JA . Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. - Proc Natl Acad Sci U S A: 1979 Apr, 76(4);1933-7 [PubMed:287033] [DOI]
- ↑ He S, Hickman AB, Varani AM, Siguier P, Chandler M, Dekker JP, Dyda F . Insertion Sequence IS26 Reorganizes Plasmids in Clinically Isolated Multidrug-Resistant Bacteria by Replicative Transposition. - mBio: 2015 Jun 9, 6(3);e00762 [PubMed:26060276] [DOI]
- ↑ Curcio MJ, Derbyshire KM . The outs and ins of transposition: from mu to kangaroo. - Nat Rev Mol Cell Biol: 2003 Nov, 4(11);865-77 [PubMed:14682279] [DOI]
- ↑ Ross DG, Swan J, Kleckner N . Nearly precise excision: a new type of DNA alteration associated with the translocatable element Tn10. - Cell: 1979 Apr, 16(4);733-8 [PubMed:455447] [DOI]
- ↑ Ohtsubo E, Zenilman M, Ohtsubo H . Plasmids containing insertion elements are potential transposons. - Proc Natl Acad Sci U S A: 1980 Feb, 77(2);750-4 [PubMed:6244582] [DOI]
- ↑ Polard P, Seroude L, Fayet O, Prère MF, Chandler M . One-ended insertion of IS911. - J Bacteriol: 1994 Feb, 176(4);1192-6 [PubMed:8106332] [DOI]