General Information/Overview
|
Intracellular MGE or transposable elements (TE) include transposons (Tn) and insertion sequences (IS) but can embrace integrons (In) [3][4] and introns [5][6][7]. Originally Tn were distinguished from IS since they carry passenger (also called cargo) genes not involved in catalyzing or regulating TE movement. Most eukaryotic DNA transposons have relatives among the prokaryotic IS (see [8]) and it is not surprising that a variety of these elements carrying passenger genes are now also being identified [9][10]. Prokaryotes harbor a host of such elements as well as several types of structure possessing characteristics of both groups (e.g. Integrative Conjugative Elements, ICE, originally called conjugative transposons, as well as other types of non-conjunctive genomic islands) [11][12][13].
|
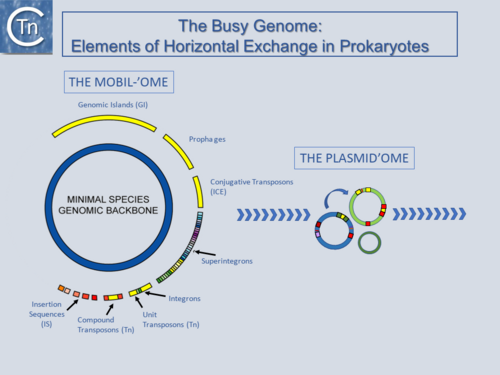 Fig.1.1.1. The Busy Genome: Elements of Horizontal Exchange. The genome backbone, which includes housekeeping genes, is shown as the inner circle (blue). The "mobilome" is shown in the outer circle. This includes a number of different types of MGE both intercellular (some genomic islands, prophages, and conjugative transposons) and intracellular (Insertion sequences, compound and unit transposons, integrons, and super integrons). An important class of intercellular MGE, the plasmids, act as transposon vectors and facilitate TE movement within the plasmidome. |
Bibliography
- ↑ Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R . The microbial pan-genome. - Curr Opin Genet Dev: 2005 Dec, 15(6);589-94 [PubMed:16185861] [DOI] </nowiki>
- ↑ Tettelin H, Riley D, Cattuto C, Medini D . Comparative genomics: the bacterial pan-genome. - Curr Opin Microbiol: 2008 Oct, 11(5);472-7 [PubMed:19086349] [DOI] </nowiki>
- ↑ Escudero JA, Loot C, Nivina A, Mazel D . The Integron: Adaptation On Demand. - Microbiol Spectr: 2015 Apr, 3(2);MDNA3-0019-2014 [PubMed:26104695] [DOI] </nowiki>
- ↑ Escudero JA, Loot C, Parissi V, Nivina A, Bouchier C, Mazel D . Unmasking the ancestral activity of integron integrases reveals a smooth evolutionary transition during functional innovation. - Nat Commun: 2016 Mar 10, 7;10937 [PubMed:26961432] [DOI] </nowiki>
- ↑ Lambowitz AM, Belfort M . Mobile Bacterial Group II Introns at the Crux of Eukaryotic Evolution. - Microbiol Spectr: 2015 Feb, 3(1);MDNA3-0050-2014 [PubMed:26104554] [DOI] </nowiki>
- ↑ Zimmerly S, Semper C . Evolution of group II introns. - Mob DNA: 2015, 6;7 [PubMed:25960782] [DOI] </nowiki>
- ↑ Zimmerly S, Wu L . An Unexplored Diversity of Reverse Transcriptases in Bacteria. - Microbiol Spectr: 2015 Apr, 3(2);MDNA3-0058-2014 [PubMed:26104699] [DOI] </nowiki>
- ↑ Hickman AB, Chandler M, Dyda F . Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. - Crit Rev Biochem Mol Biol: 2010 Feb, 45(1);50-69 [PubMed:20067338] [DOI] </nowiki>
- ↑ Bao W, Jurka MG, Kapitonov VV, Jurka J . New superfamilies of eukaryotic DNA transposons and their internal divisions. - Mol Biol Evol: 2009 May, 26(5);983-93 [PubMed:19174482] [DOI] </nowiki>
- ↑ Bao W, Jurka J . Homologues of bacterial TnpB_IS605 are widespread in diverse eukaryotic transposable elements. - Mob DNA: 2013 Apr 1, 4(1);12 [PubMed:23548000] [DOI] </nowiki>
- ↑ Burrus V, Waldor MK . Shaping bacterial genomes with integrative and conjugative elements. - Res Microbiol: 2004 Jun, 155(5);376-86 [PubMed:15207870] [DOI] </nowiki>
- ↑ Dobrindt U, Hochhut B, Hentschel U, Hacker J . Genomic islands in pathogenic and environmental microorganisms. - Nat Rev Microbiol: 2004 May, 2(5);414-24 [PubMed:15100694] [DOI] </nowiki>
- ↑ Guérillot R, Da Cunha V, Sauvage E, Bouchier C, Glaser P . Modular evolution of TnGBSs, a new family of integrative and conjugative elements associating insertion sequence transposition, plasmid replication, and conjugation for their spreading. - J Bacteriol: 2013 May, 195(9);1979-90 [PubMed:23435978] [DOI] </nowiki>